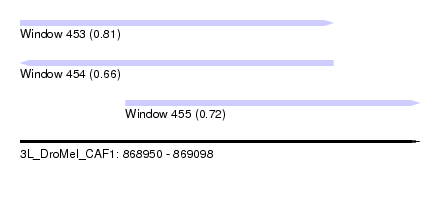
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 868,950 – 869,098 |
| Length | 148 |
| Max. P | 0.814109 |

| Location | 868,950 – 869,066 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -15.82 |
| Energy contribution | -17.30 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 868950 116 + 23771897 AUUCAUUCGGUGAAUGUAUAAAACCAUGUAUAUC-AAGAAGUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UAUUGCUAUCUAUAUGUGUCAUAAUAGCAUAUACUGCAG .....(((..(((.((((((......))))))))-).))).(((((..(.(((...(((...((((((((.(((((..-..))))).)))...)))))...))).))).)..))))). ( -25.20) >DroSec_CAF1 11873 114 + 1 AU--AUGUGGUGAAUGUAUAAAUCCAUGUAUAUC-GAGAAGUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UCUUGCUAUCUAUAUGUGCCAUACUUGCAUAUACUGCUG ((--(((..((((((((((((((...(((((...-.....)))))..)))))))).))))).((((((((.(((((..-..))))).)))...))))).....)..)))))....... ( -27.50) >DroSim_CAF1 6894 114 + 1 AU--AUGUGGUGAAUGUAUAAAUCCAUGUAUAUC-GAGAAGUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UCUUGCUAUCUAUAUGUGCCAUAAUUGCAUAUACUGCUG ((--(((..((...(((((((((...(((((...-.....)))))..)))))))))......((((((((.(((((..-..))))).)))...)))))....))..)))))....... ( -28.80) >DroEre_CAF1 11902 107 + 1 AU--AUUGUGUGGAUGUAUAAAACCUUGAAUACC-AAGAAGUGCAACGUUUGUAUAAUUCAAGCACAAGAAAGAAACUUU--------CUAUAUGUGCCAUAAUUGCAUAUACUGCAG ..--((((((..(((((.......((((.....)-))).......)))))..))))))....(((((((((((...))))--------))...)))))......((((.....)))). ( -23.54) >DroYak_CAF1 11941 110 + 1 UU--AUUGC------AUAUAAAUGCAUGAAUAUUGAAAAAACGCAGCGUUUGUAUAAUUCAAGCACAAGAAAGAAACUUUCUUGCUAUCUAUAUGUGCCAUAAUUGCAUACACUGCAG ((--((.((------(((((..(((.(((((..((...(((((...))))).))..))))).)))((((((((...)))))))).......))))))).)))).((((.....)))). ( -26.30) >consensus AU__AUUGGGUGAAUGUAUAAAUCCAUGUAUAUC_AAGAAGUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU_UCUUGCUAUCUAUAUGUGCCAUAAUUGCAUAUACUGCAG ..........................................((((.((.((((........((((((((.(((((.....))))).)))...)))))......)))).)).)))).. (-15.82 = -17.30 + 1.48)

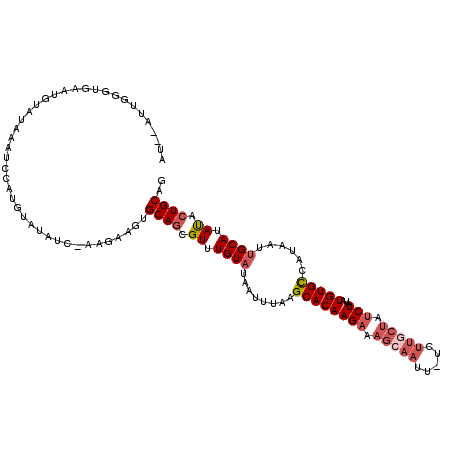
| Location | 868,950 – 869,066 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -14.11 |
| Energy contribution | -15.55 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 868950 116 - 23771897 CUGCAGUAUAUGCUAUUAUGACACAUAUAGAUAGCAAUA-AAUUGCUUUCUUGUGCUUAAAUUAUACAAACGCUGCACUUCUU-GAUAUACAUGGUUUUAUACAUUCACCGAAUGAAU .(((((..(.((.(((..(((((((...(((.(((((..-..))))).))))))).)))....))))).)..)))))......-..................(((((...)))))... ( -20.10) >DroSec_CAF1 11873 114 - 1 CAGCAGUAUAUGCAAGUAUGGCACAUAUAGAUAGCAAGA-AAUUGCUUUCUUGUGCUUAAAUUAUACAAACGCUGCACUUCUC-GAUAUACAUGGAUUUAUACAUUCACCACAU--AU ..((((..(.((..(((..((((((...(((.(((((..-..))))).)))))))))...)))...)).)..)))).......-........(((((......)))))......--.. ( -24.90) >DroSim_CAF1 6894 114 - 1 CAGCAGUAUAUGCAAUUAUGGCACAUAUAGAUAGCAAGA-AAUUGCUUUCUUGUGCUUAAAUUAUACAAACGCUGCACUUCUC-GAUAUACAUGGAUUUAUACAUUCACCACAU--AU ..((((..(.((.((((..((((((...(((.(((((..-..))))).)))))))))..))))...)).)..)))).......-........(((((......)))))......--.. ( -25.50) >DroEre_CAF1 11902 107 - 1 CUGCAGUAUAUGCAAUUAUGGCACAUAUAG--------AAAGUUUCUUUCUUGUGCUUGAAUUAUACAAACGUUGCACUUCUU-GGUAUUCAAGGUUUUAUACAUCCACACAAU--AU .....(((((((((((...((((((...((--------((((...))))))))))))..............))))))..((((-(.....)))))...)))))...........--.. ( -19.83) >DroYak_CAF1 11941 110 - 1 CUGCAGUGUAUGCAAUUAUGGCACAUAUAGAUAGCAAGAAAGUUUCUUUCUUGUGCUUGAAUUAUACAAACGCUGCGUUUUUUCAAUAUUCAUGCAUUUAUAU------GCAAU--AA ..(((((((.((.((((..((((((...(((.(((......))))))....))))))..))))...)).)))))))................(((((....))------)))..--.. ( -27.10) >consensus CUGCAGUAUAUGCAAUUAUGGCACAUAUAGAUAGCAAGA_AAUUGCUUUCUUGUGCUUAAAUUAUACAAACGCUGCACUUCUU_GAUAUACAUGGAUUUAUACAUUCACCAAAU__AU ..((((..(.((.......((((((...(((.(((((.....))))).))))))))).........)).)..)))).......................................... (-14.11 = -15.55 + 1.44)

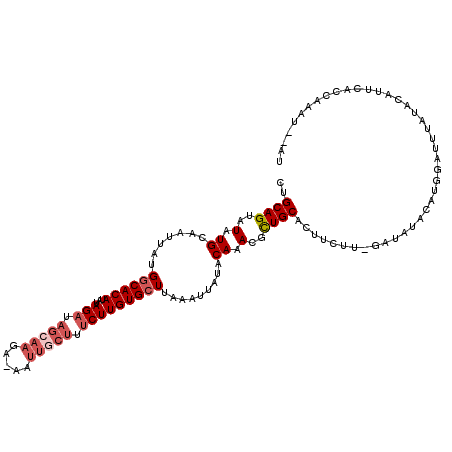
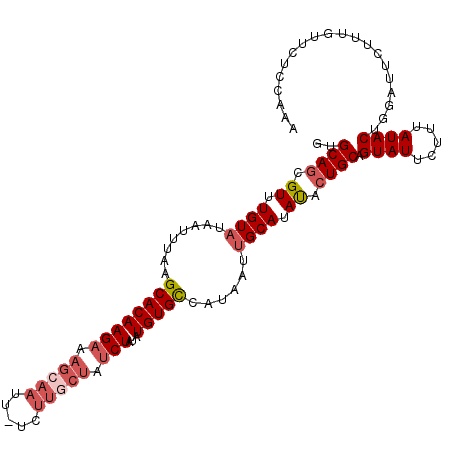
| Location | 868,989 – 869,098 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -16.94 |
| Energy contribution | -18.42 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 868989 109 + 23771897 GUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UAUUGCUAUCUAUAUGUGUCAUAAUAGCAUAUACUGCAGUAUUCUUUAUACUGGAUUCUUUGUUCUCCAAA .(((((..(.(((...(((...((((((((.(((((..-..))))).)))...)))))...))).))).)..)))))((((.....))))((((..........)))).. ( -22.90) >DroSec_CAF1 11910 109 + 1 GUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UCUUGCUAUCUAUAUGUGCCAUACUUGCAUAUACUGCUGUAUUCUUUAUACUGGAUUCUUUGUUCUCCAAA ((((((((((((.......))))).(((((((....))-))))).......(((((((.......)))))))..))))))).........((((..........)))).. ( -25.70) >DroSim_CAF1 6931 109 + 1 GUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU-UCUUGCUAUCUAUAUGUGCCAUAAUUGCAUAUACUGCUGUAUUCUUUAUACUGGAUUCUUUGUUCUCCAAA (((((((((.(((.(((((...((((((((.(((((..-..))))).)))...)))))...))))).))).)).))))))).........((((..........)))).. ( -27.60) >DroEre_CAF1 11939 102 + 1 GUGCAACGUUUGUAUAAUUCAAGCACAAGAAAGAAACUUU--------CUAUAUGUGCCAUAAUUGCAUAUACUGCAGUAUUCUUUAUACUGGCUUCUUUGUUCUCAAAA .......((..((((((.....(((((((((((...))))--------))...)))))....((((((.....)))))).....))))))..))...((((....)))). ( -20.60) >DroYak_CAF1 11973 110 + 1 ACGCAGCGUUUGUAUAAUUCAAGCACAAGAAAGAAACUUUCUUGCUAUCUAUAUGUGCCAUAAUUGCAUACACUGCAGUAUUCUUUAUACUGGUUUCUUUGUUCUCCAAA ..((((.((.((((........(((((((((((...))))))...........)))))......)))).)).))))(((((.....)))))................... ( -21.34) >consensus GUGCAGCGUUUGUAUAAUUUAAGCACAAGAAAGCAAUU_UCUUGCUAUCUAUAUGUGCCAUAAUUGCAUAUACUGCAGUAUUCUUUAUACUGGAUUCUUUGUUCUCCAAA ..((((.((.((((........((((((((.(((((.....))))).)))...)))))......)))).)).)))).((((.....)))).................... (-16.94 = -18.42 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:46 2006