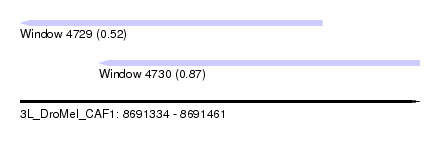
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,691,334 – 8,691,461 |
| Length | 127 |
| Max. P | 0.867457 |

| Location | 8,691,334 – 8,691,430 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8691334 96 - 23771897 CCCUAAAAUCCGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGAGGGCGC-------AAAAAG--------CGUACGAGAAAUGC ........((((.(((((.....(((.(((((((......))))))).))).(((........)))))))).((((-------.....)--------))).)).))..... ( -27.80) >DroSec_CAF1 79161 95 - 1 CCCAAAAAUCCGACUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAA-AAAGGGCAGAAGGCGA-------AAAAAU--------CGUACGAGGAAUGC ........((((.((((..(((((((((((((((......)))))))..))).))))).-..))))).....((((-------.....)--------)))....))).... ( -28.00) >DroSim_CAF1 79666 96 - 1 CCCAAAAAUCCGACUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGAAGGCGC-------AAAAAG--------CGUACGAGAAAUGC ........((((.((((..(((((((((((((((......)))))))..))).)))))....))))......((((-------.....)--------))).)).))..... ( -27.10) >DroEre_CAF1 83544 94 - 1 CGCAAAAAUCUGAUUCUGAUUCGGUGGCAUGUGCAUUGAAGCACGUGCUACACCCGAAA-AAAGAGCAGGAGGCGC-------AAAAAG--------UGUACGA-AAAUGC .(((....((((.((((..(((((((((((((((......))))))))))...))))).-..))))))))..((((-------.....)--------)))....-...))) ( -32.70) >DroYak_CAF1 81918 96 - 1 CCCUAAAAUCAGAUUCUGAUUCGGUGUCAUGUGCAUUGGAGCACGUGCUACACCCGAAAAAAAGAGCAGGCGGCGC-------AAAAAG--------UGUUCGAGAAAUGC ......((((((...)))))).((((((((((((......)))))))..)))))...........(((..((((((-------.....)--------)).))).....))) ( -25.00) >DroAna_CAF1 76002 106 - 1 --CAAAAAUCCGAACCAGAUUUGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAGAA---CACAAGACCCCAACAAAAAAAAGUGUAAGAUUGUACGAAAAAUGC --........((.((..(((((((((((((((((......)))))))..)))))........(---(((...................)))).))))))).))........ ( -19.11) >consensus CCCAAAAAUCCGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGAAGGCGC_______AAAAAG________CGUACGAGAAAUGC ...........(.((((..(((((((((((((((......)))))))..))).)))))....)))))............................................ (-17.49 = -17.85 + 0.36)

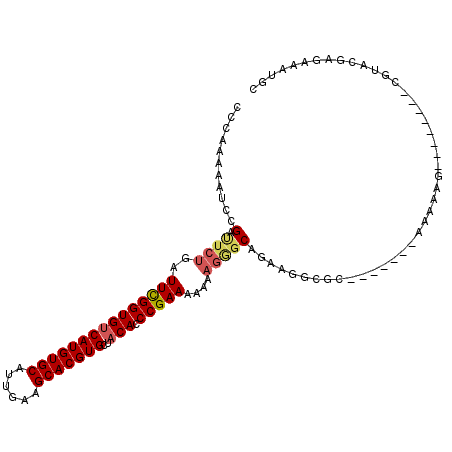
| Location | 8,691,359 – 8,691,461 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
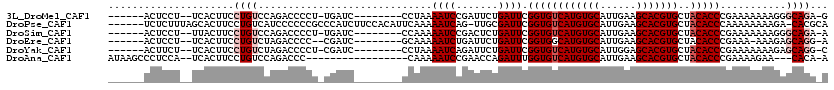
>3L_DroMel_CAF1 8691359 102 - 23771897 ------ACUCCU--UCACUUCCUGUCCAGACCCCU-UGAUC--------CCUAAAAUCCGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGA-G ------......--.......((((((......(.-.((((--------..........))))..).(((((((((((((((......)))))))..))).))))).....)))))).-. ( -27.60) >DroPse_CAF1 85220 112 - 1 ------UCUCUUUAGCACUUCCUGUCAUCCCCCCGCCCAUCUUCCACAUUCAAAAAUCAG-UUGCGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCAAAAAAAAGA-CACGCA ------................((((.......(((........................-..)))....((((((((((((......)))))))..))))).........))-)).... ( -20.87) >DroSim_CAF1 79691 102 - 1 ------ACUCCU--UUACUUCCUGUCCAGACCCCU-UGAUC--------CCAAAAAUCCGACUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGGGCAGA-A ------......--.......((((((.......(-((...--------.)))..............(((((((((((((((......)))))))..))).))))).....)))))).-. ( -27.20) >DroEre_CAF1 83568 100 - 1 ------ACUCCU--UCACUUCCUGUCUAGACCCC--CGAUC--------GCAAAAAUCUGAUUCUGAUUCGGUGGCAUGUGCAUUGAAGCACGUGCUACACCCGAAA-AAAGAGCAGG-A ------......--.....((((((((.......--.((((--------(........)))))....(((((((((((((((......))))))))))...))))).-..))).))))-) ( -30.20) >DroYak_CAF1 81943 102 - 1 ------ACUUCU--UCACUUCCUGUCUAGACCCCU-CGAUC--------CCUAAAAUCAGAUUCUGAUUCGGUGUCAUGUGCAUUGGAGCACGUGCUACACCCGAAAAAAAGAGCAGG-C ------......--......((((.((.......(-((...--------.....((((((...)))))).((((((((((((......)))))))..)))))))).......))))))-. ( -27.24) >DroAna_CAF1 76042 97 - 1 AUAAGCCCUCCA--UCACUUCCUGUCCAGACCC-----------------CAAAAAUCCGAACCAGAUUUGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAGAA---CACA-A ............--((..(((............-----------------...(((((.......)))))((((((((((((......)))))))..))))).)))..)).---....-. ( -17.80) >consensus ______ACUCCU__UCACUUCCUGUCCAGACCCCU_CGAUC________CCAAAAAUCCGAUUCUGAUUCGGUGUCAUGUGCAUUGAAGCACGUGCUACACCCGAAAAAAAGAGCAGA_A .....................((((.............................((((.......)))).((((((((((((......)))))))..)))))...........))))... (-15.97 = -16.80 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:51 2006