
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,687,668 – 8,687,913 |
| Length | 245 |
| Max. P | 0.999087 |

| Location | 8,687,668 – 8,687,783 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
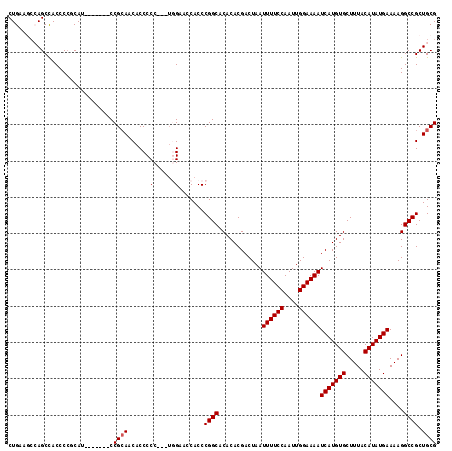
>3L_DroMel_CAF1 8687668 115 + 23771897 CUGAAGCCAGCCACCCGCCAUCCGCUAUCCGCAACACCCCC---UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG ..(.(((..(((....(((....((.....)).........---.((......)))))............((((((....))))))(((((((.....)))))))...))).))).). ( -27.10) >DroSec_CAF1 75459 108 + 1 CUGAAGACAGCCACCCCGCAU-------CCGAAACACCCCC---UGGAGCCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG .....(.((((......((.(-------(((..........---)))))).....(((............((((((....))))))(((((((.....)))))))....)))))))). ( -26.50) >DroSim_CAF1 75937 108 + 1 CUGAAGCCAGCUACCCCUCAU-------CCGCAACACCCCC---UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG .....((.(((.........(-------(((..........---)))).......(((............((((((....))))))(((((((.....)))))))....)))))))). ( -24.40) >DroEre_CAF1 79823 108 + 1 CUGAACCGAGCCACCCCGCAU-------CCGCAACACCCCC---UCGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG ......((((.......((..-------..))........)---))).......((((............((((((....))))))(((((((.....)))))))....))))..... ( -22.26) >DroYak_CAF1 78089 111 + 1 CUGAACCAAGCCACCCCUCAU-------CCGCAACACCCCCCGCGCGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG .........((.........(-------((((..........))).))......((((............((((((....))))))(((((((.....)))))))....))))..)). ( -22.90) >consensus CUGAAGCCAGCCACCCCGCAU_______CCGCAACACCCCC___UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCG .............................((((......(......).......((((............((((((....))))))(((((((.....)))))))....)))).)))) (-18.90 = -19.10 + 0.20)


| Location | 8,687,668 – 8,687,783 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687668 115 - 23771897 CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA---GGGGGUGUUGCGGAUAGCGGAUGGCGGGUGGCUGGCUUCAG ..((((.((((..................(((((((((....)))))))))(((((.(((.(((.(..(..(..---..)..)..).)))...))).))))))))).))))....... ( -38.80) >DroSec_CAF1 75459 108 - 1 CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGCUCCA---GGGGGUGUUUCGG-------AUGCGGGGUGGCUGUCUUCAG .(((((.(((((..(((.......((((((((((((((....)))))....))))))))).(((((..((((..---..))))..)))))-------))).))))).)))))...... ( -41.10) >DroSim_CAF1 75937 108 - 1 CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA---GGGGGUGUUGCGG-------AUGAGGGGUAGCUGGCUUCAG ..((((.((((((.(((.(((((.((((((((((((((....)))))....))))))))).(((((....))).---)).....))))).-------))))))))).))))....... ( -37.60) >DroEre_CAF1 79823 108 - 1 CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCGA---GGGGGUGUUGCGG-------AUGCGGGGUGGCUCGGUUCAG (((((((.(((((((.........((((((((((((((....)))))....)))))))))(((....)))..))---))))))))))))(-------(..((((....))))..)).. ( -39.60) >DroYak_CAF1 78089 111 - 1 CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCGCGCGGGGGGUGUUGCGG-------AUGAGGGGUGGCUUGGUUCAG ((.(((.((((((.(((.((((..((((.(((((((((....))))))))).(((((((.(((....))).)))))))...)))))))).-------))))))))).)))))...... ( -41.80) >consensus CGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA___GGGGGUGUUGCGG_______AUGAGGGGUGGCUGGCUUCAG (((((((.((((............((((((((((((((....)))))....)))))))))(((....))).......)))).)))))))............................. (-31.20 = -31.24 + 0.04)



| Location | 8,687,706 – 8,687,823 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -35.80 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
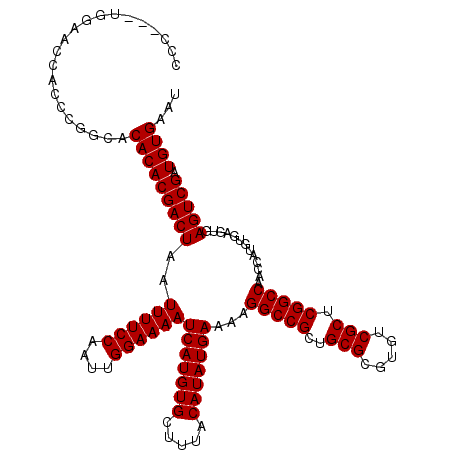
>3L_DroMel_CAF1 8687706 117 + 23771897 CCC---UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU ..(---(((......))))..(((((((((....(((((..(((....(((((((.....)))))))...(((((..(((.....))).)))))..))))).)))..))))).))))... ( -39.40) >DroSec_CAF1 75490 117 + 1 CCC---UGGAGCCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU ...---....(((....))).(((((((((....(((((..(((....(((((((.....)))))))...(((((..(((.....))).)))))..))))).)))..))))).))))... ( -40.30) >DroSim_CAF1 75968 117 + 1 CCC---UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU ..(---(((......))))..(((((((((....(((((..(((....(((((((.....)))))))...(((((..(((.....))).)))))..))))).)))..))))).))))... ( -39.40) >DroEre_CAF1 79854 117 + 1 CCC---UCGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU ...---((((..(((..((((((...((....((((((....)))))))).))))))...((((((....(((((..(((.....))).)))))...)))))).)))..))))....... ( -36.50) >DroYak_CAF1 78120 120 + 1 CCCCGCGCGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU ...(((((((..(((..((((((...((....((((((....)))))))).))))))...((((((....(((((..(((.....))).)))))...)))))).)))..))).))))... ( -39.30) >DroAna_CAF1 72652 107 + 1 -------------CCCCACCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUAUGAGUGAGUCGAUGUGAAU -------------........(((((((((..((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).............))))).))))... ( -35.80) >consensus CCC___UGGAACCACCCGGCACACACGACUAAUUUUCCAAUUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAU .....................(((((((((..((((((....))))))(((((((.....)))))))...(((((..(((.....))).))))).............))))).))))... (-35.80 = -35.80 + 0.00)


| Location | 8,687,706 – 8,687,823 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -37.21 |
| Energy contribution | -37.90 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687706 117 - 23771897 AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA---GGG .........((..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))..))..---... ( -39.70) >DroSec_CAF1 75490 117 - 1 AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGCUCCA---GGG .........((..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))..))..---... ( -39.70) >DroSim_CAF1 75968 117 - 1 AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA---GGG .........((..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))..))..---... ( -39.70) >DroEre_CAF1 79854 117 - 1 AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCGA---GGG .....(.((((..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))..))))---.). ( -44.10) >DroYak_CAF1 78120 120 - 1 AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCGCGCGGGG ..((....(((..((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))..)))....)). ( -41.80) >DroAna_CAF1 72652 107 - 1 AUUCACAUCGACUCACUCAUAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGGUGGGG------------- ...........(((((((((((((..(((((.(((.....)))..)))))...)))))))....((((((((((((((....)))))....))))))))))))))).------------- ( -42.20) >consensus AUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAAUUGGAAAAUUAGUCGUGUGUGCCGGGUGGUUCCA___GGG .........((.(((((((((((...(((((.(((.....)))..)))))......)))))...((((((((((((((....)))))....)))))))))...)))))).))........ (-37.21 = -37.90 + 0.69)



| Location | 8,687,743 – 8,687,862 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -32.25 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
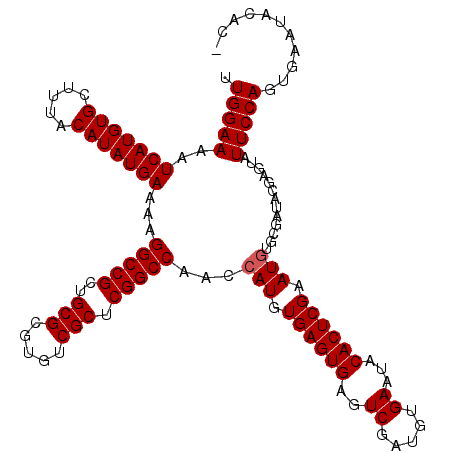
>3L_DroMel_CAF1 8687743 119 + 23771897 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACAC- .(((....(((((((.....)))))))...(((((..(((.....))).)))))..)))((((.(((..((((.(((....)))))))...)))..)))).((((((....))))))..- ( -36.80) >DroSec_CAF1 75527 119 + 1 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACAC- .(((....(((((((.....)))))))...(((((..(((.....))).)))))..)))((((.(((..((((.(((....)))))))...)))..)))).((((((....))))))..- ( -36.80) >DroSim_CAF1 76005 119 + 1 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACAC- .(((....(((((((.....)))))))...(((((..(((.....))).)))))..)))((((.(((..((((.(((....)))))))...)))..)))).((((((....))))))..- ( -36.80) >DroEre_CAF1 79891 117 + 1 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUA--C- ((((((..(((((((.....))))))).........(((.((((((((.(((....))...((((((..((.....))...))))))...).)))))))).)))))))))......--.- ( -35.50) >DroYak_CAF1 78160 117 + 1 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCACUGAAUA--C- .(((....(((((((.....)))))))...(((((..(((.....))).)))))..)))((((.(((..((((.(((....)))))))...)))..)))).((((((....)))))--)- ( -35.10) >DroAna_CAF1 72679 109 + 1 UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUAUGAGUGAGUCGAUGUGAAUACACUCGAAUA-----------UAUUCCACUGAAUAUACU .(((....(((((((.....)))))))...(((((..(((.....))).)))))..)))..((((((..((.....))...))))))..((-----------(((((....))))))).. ( -32.80) >consensus UUGGAAAAUCAUGUGCUUUACAUAUGAAAAGGCCGCUGCGCGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACAC_ .(((((..(((((((.....)))))))...(((((..(((.....))).)))))...(((.((((((..((.....))...)))))).))).............)))))........... (-32.25 = -32.42 + 0.17)

| Location | 8,687,743 – 8,687,862 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -28.50 |
| Energy contribution | -29.37 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
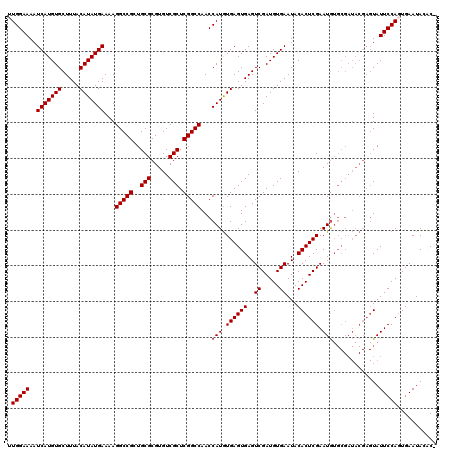
>3L_DroMel_CAF1 8687743 119 - 23771897 -GUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA -(((....)))(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). ( -33.10) >DroSec_CAF1 75527 119 - 1 -GUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA -(((....)))(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). ( -33.10) >DroSim_CAF1 76005 119 - 1 -GUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA -(((....)))(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). ( -33.10) >DroEre_CAF1 79891 117 - 1 -G--UAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA -.--.......(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). ( -31.70) >DroYak_CAF1 78160 117 - 1 -G--UAUUCAGUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA -.--.......(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). ( -31.80) >DroAna_CAF1 72679 109 - 1 AGUAUAUUCAGUGGAAUA-----------UAUUCGAGUGUAUUCACAUCGACUCACUCAUAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA .((((((((....)))))-----------)))(((((((....))).))))......(((((((..(((((.(((.....)))..)))))...))))))).................... ( -31.20) >consensus _GUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACGCGCAGCGGCCUUUUCAUAUGUAAAGCACAUGAUUUUCCAA ...........(((((...((((...((......(((((...((.....))..)))))(((((...(((((.(((.....)))..)))))......)))))...))..))))..))))). (-28.50 = -29.37 + 0.86)



| Location | 8,687,783 – 8,687,876 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687783 93 + 23771897 CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUA----------UG-----GUA ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).((((((....))))))((((.......----------))-----)). ( -28.50) >DroSec_CAF1 75567 93 + 1 CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUA----------UG-----GUA ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).((((((....))))))((((.......----------))-----)). ( -28.50) >DroSim_CAF1 76045 93 + 1 CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUA----------UG-----GUA ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).((((((....))))))((((.......----------))-----)). ( -28.50) >DroEre_CAF1 79931 86 + 1 CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUA--CCACUGUAUA-------------------- ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).......(((((....--.)))))....-------------------- ( -26.50) >DroYak_CAF1 78200 106 + 1 CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCACUGAAUA--CCACCGUAUACCGCCGUAUACCGCCGUGUA ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).((((((....)))))--)(((.((((((....))))))....))).. ( -30.60) >consensus CGUGUCGCUCGGCCAACCAUGUGAGUGAGUCGAUGUGAAUACACUCGAAUGUGCGAUACGAGUAUUCCAGUGAAUACACCACUGUAUA__________UG_____GUA ((((((((.(((....))...((((((..((.....))...))))))...).)))))))).......(((((.......)))))........................ (-24.52 = -24.92 + 0.40)


| Location | 8,687,783 – 8,687,876 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687783 93 - 23771897 UAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG .((-----((----------(.....)))))((((((....)))))).(((.((((......(((((...((.....))..)))))...(((....))).)))).))) ( -26.40) >DroSec_CAF1 75567 93 - 1 UAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG .((-----((----------(.....)))))((((((....)))))).(((.((((......(((((...((.....))..)))))...(((....))).)))).))) ( -26.40) >DroSim_CAF1 76045 93 - 1 UAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG .((-----((----------(.....)))))((((((....)))))).(((.((((......(((((...((.....))..)))))...(((....))).)))).))) ( -26.40) >DroEre_CAF1 79931 86 - 1 --------------------UAUACAGUGG--UAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG --------------------....(((((.--....))))).......(((.((((......(((((...((.....))..)))))...(((....))).)))).))) ( -23.10) >DroYak_CAF1 78200 106 - 1 UACACGGCGGUAUACGGCGGUAUACGGUGG--UAUUCAGUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG ......((.((((((....)))))).))((--(((((....)))))))(((.((((......(((((...((.....))..)))))...(((....))).)))).))) ( -31.40) >consensus UAC_____CA__________UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGUAUUCACAUCGACUCACUCACAUGGUUGGCCGAGCGACACG ........................((((((.....)))))).......(((.((((......(((((...((.....))..)))))...(((....))).)))).))) (-23.02 = -23.06 + 0.04)


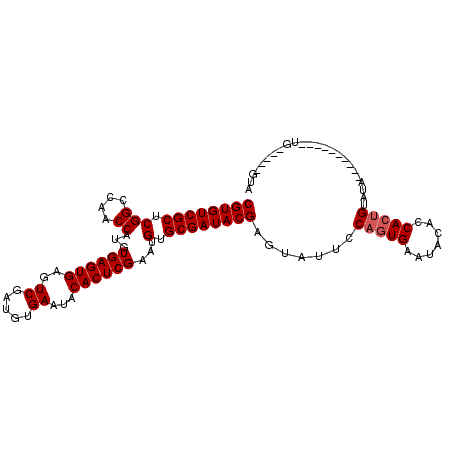
| Location | 8,687,823 – 8,687,913 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687823 90 - 23771897 GAUAUAAUUGCGUUUUUUCGUGAAUGUAUAC--AGCAUAUAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGU .(((((.(..((......))..).))))).(--((.((((((-----((----------(.....)))))))))...)))..(((((((...........))))))) ( -23.60) >DroSec_CAF1 75607 90 - 1 GAUAUAAUUGCGUUUUUUCGUGAAUGUAUAC--AGCGUAUAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGU .(((((.(..((......))..).))))).(--((.((((((-----((----------(.....)))))))))...)))..(((((((...........))))))) ( -23.90) >DroSim_CAF1 76085 90 - 1 GAUAUAAUUGCGUUUUUUCGUGAAUGUAUAC--AGCGUAUAC-----CA----------UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGU .(((((.(..((......))..).))))).(--((.((((((-----((----------(.....)))))))))...)))..(((((((...........))))))) ( -23.90) >DroEre_CAF1 79971 80 - 1 GAUAUAAUUGCGUUU-UUCGUGAAUGUAUAC--AGCA----------------------UAUACAGUGG--UAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGU .(((((.(..((...-..))..).)))))..--....----------------------....(((((.--....)))))..(((((((...........))))))) ( -18.00) >DroYak_CAF1 78240 105 - 1 GAUAUAAUUGCGUUUUUUCGUGAAUGUAUACACAGCACAUACACGGCGGUAUACGGCGGUAUACGGUGG--UAUUCAGUGGAAUACUCGUAUCGCACAUUCGAGUGU (((((.............((((.((((.........)))).))))((.((((((....)))))).))((--(((((....))))))).)))))((((......)))) ( -26.60) >consensus GAUAUAAUUGCGUUUUUUCGUGAAUGUAUAC__AGCAUAUAC_____CA__________UAUACAGUGGUGUAUUCACUGGAAUACUCGUAUCGCACAUUCGAGUGU .(((((.(..((......))..).)))))..................................((((((.....))))))..(((((((...........))))))) (-18.12 = -18.16 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:47 2006