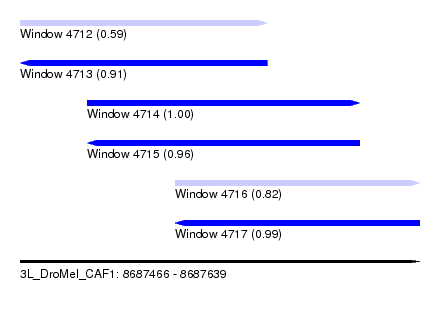
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,687,466 – 8,687,639 |
| Length | 173 |
| Max. P | 0.998221 |

| Location | 8,687,466 – 8,687,573 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -23.04 |
| Energy contribution | -24.24 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687466 107 + 23771897 CGGACCUCUUUU--AUUCGGCGGGCUUUGCCAAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCUCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCU ............--....(((((((((((((...-...)))))).)))))))..........(((((....(((.((...(((.....)))....)))))..)))))... ( -35.30) >DroSec_CAF1 75253 109 + 1 UGGACCUCUUUUUUUUUUGGCGGGCUUAGUCAAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCU ..((((............(((((((((.(((...-...))).(((((((..............)))))))))))))))).(((.....))).....))))(((...))). ( -30.54) >DroSim_CAF1 75732 109 + 1 UGGACCUCUUUUUUUUUUGGCGGGCUUAGUCAAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCU ..((((............(((((((((.(((...-...))).(((((((..............)))))))))))))))).(((.....))).....))))(((...))). ( -30.54) >DroEre_CAF1 79625 98 + 1 C-----------UUUUUCGGCGGUCUUGCCCAAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCU .-----------......((((((.(((((....-...)))))..)))...........(((((((.((((((((.........)))))))))))))))...)))..... ( -27.10) >DroAna_CAF1 72420 108 + 1 UGGGCUUUUUGUAUUUUU--AGAGCUUAGCGAAUUUUCGGCGAAUGCCUGUCUAUAAUUACUCAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUUGGUUGCGCCUCGCU ((((((((..(....)..--))))))))((((......((((((((((((..((....))..)))))))).((((.....(((.....))).....)))).)))))))). ( -33.80) >consensus UGGACCUCUUUUUUUUUUGGCGGGCUUAGCCAAU_UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCU ..................(((((((...(((.......)))....)))))))..........(((((....((((.....(((.....))).....))))..)))))... (-23.04 = -24.24 + 1.20)


| Location | 8,687,466 – 8,687,573 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.14 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687466 107 - 23771897 AGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGAGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUUGGCAAAGCCCGCCGAAU--AAAAGAGGUCCG .(((.(((......(((....((((..((....)).))))..(((((..((........))))))))))((((..-..))))...)))))).....--............ ( -26.80) >DroSec_CAF1 75253 109 - 1 AGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUUGACUAAGCCCGCCAAAAAAAAAAGAGGUCCA .(((.(((.....(((.....(((((.((....)))))))(((((((..((........))))))))).......-.))).....))))))................... ( -26.60) >DroSim_CAF1 75732 109 - 1 AGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUUGACUAAGCCCGCCAAAAAAAAAAGAGGUCCA .(((.(((.....(((.....(((((.((....)))))))(((((((..((........))))))))).......-.))).....))))))................... ( -26.60) >DroEre_CAF1 79625 98 - 1 AGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUUGGGCAAGACCGCCGAAAAA-----------G .....((((............(((((.((....)))))))(((((((..((........)))))))))))))...-.((.(((......))).))...-----------. ( -28.50) >DroAna_CAF1 72420 108 - 1 AGCGAGGCGCAACCAAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUGAGUAAUUAUAGACAGGCAUUCGCCGAAAAUUCGCUAAGCUCU--AAAAAUACAAAAAGCCCA (((((((((............(((((.((....)))))))((((((((..((....))..))))))))))))......))))).......--.................. ( -29.70) >consensus AGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA_AUUGGCUAAGCCCGCCAAAAAAAAAAGAGGUCCA .(((.(((....((((.....(((((.((....)))))))(((((((..((........))))))))).........))))....))))))................... (-22.06 = -23.14 + 1.08)

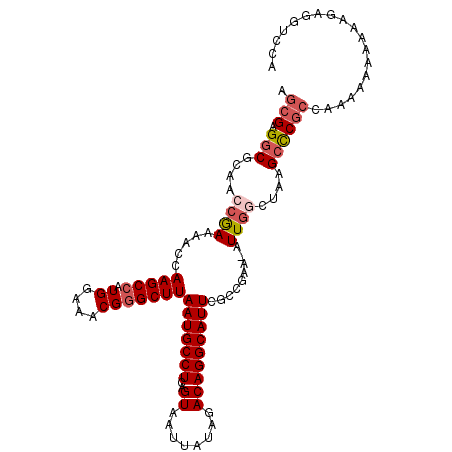
| Location | 8,687,495 – 8,687,613 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.87 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -40.01 |
| Energy contribution | -39.90 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687495 118 + 23771897 AAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCUCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG ...-...(((((..(((.(((((..((....((((((((((((.(((....))))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... ( -40.20) >DroSec_CAF1 75284 118 + 1 AAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG ...-...(((((..(((.(((((..((....(((((((((((((.........))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... ( -41.10) >DroSim_CAF1 75763 118 + 1 AAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG ...-...(((((..(((.(((((..((....(((((((((((((.........))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... ( -41.10) >DroEre_CAF1 79645 118 + 1 AAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG ...-...(((((..(((.(((((..((....(((((((((((((.........))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... ( -41.10) >DroYak_CAF1 77895 118 + 1 AAU-UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG ...-...(((((..(((.(((((..((....(((((((((((((.........))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... ( -41.10) >DroAna_CAF1 72449 119 + 1 AAUUUUCGGCGAAUGCCUGUCUAUAAUUACUCAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUUGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG .......((((((((((((..((....))..)))))))).((((.....(((.....))).....)))).))))..((.(((((((((..........))))))))))).......... ( -42.30) >consensus AAU_UUCGGCGAAUGCCUGUCUAUAAUUACUGAGGCAUUAAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACG .......(((((..(((.(((((..((....(((((((((((((.........))))))))...((((((((...))).))))))))))....))..))))).)))....))))).... (-40.01 = -39.90 + -0.11)

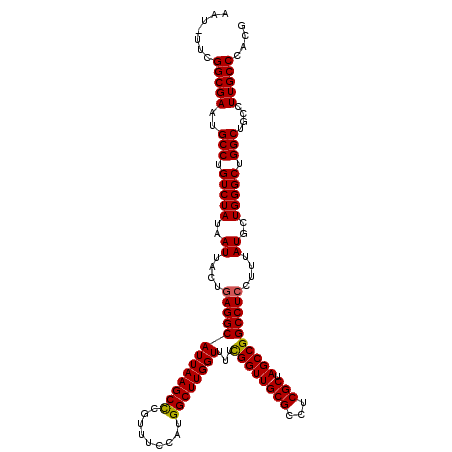
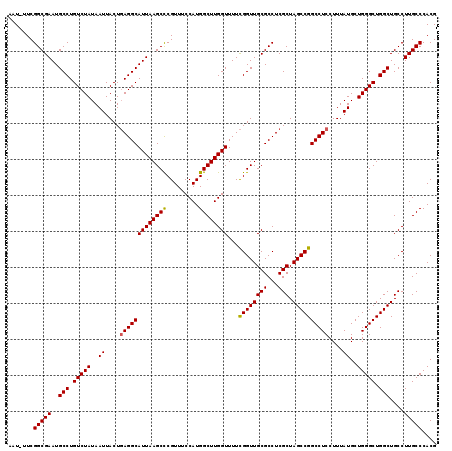
| Location | 8,687,495 – 8,687,613 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.87 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -35.65 |
| Energy contribution | -35.82 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687495 118 - 23771897 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGAGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUU (((.(((..(((..((.............))..))).))).))).((((............((((..((....)).))))(((((((..((........)))))))))))))...-... ( -32.22) >DroSec_CAF1 75284 118 - 1 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))(((((((..((........)))))))))))))...-... ( -36.72) >DroSim_CAF1 75763 118 - 1 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))(((((((..((........)))))))))))))...-... ( -36.72) >DroEre_CAF1 79645 118 - 1 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))(((((((..((........)))))))))))))...-... ( -36.72) >DroYak_CAF1 77895 118 - 1 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA-AUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))(((((((..((........)))))))))))))...-... ( -36.72) >DroAna_CAF1 72449 119 - 1 CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCAAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUGAGUAAUUAUAGACAGGCAUUCGCCGAAAAUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))((((((((..((....))..))))))))))))....... ( -39.52) >consensus CGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUUAAUGCCUCAGUAAUUAUAGACAGGCAUUCGCCGAA_AUU (((.(((..(((..((.............))..))).))).))).((((............(((((.((....)))))))(((((((..((........)))))))))))))....... (-35.65 = -35.82 + 0.17)

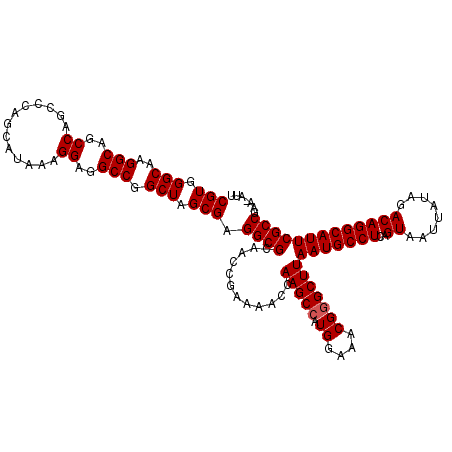
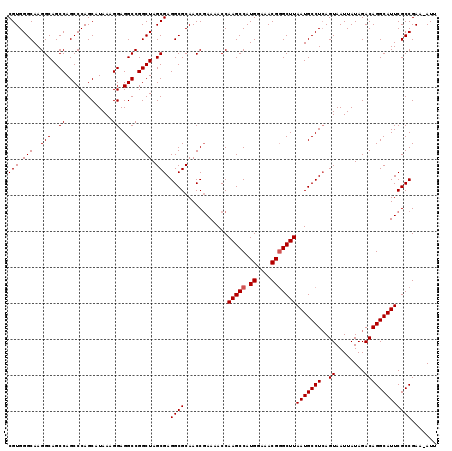
| Location | 8,687,533 – 8,687,639 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -38.48 |
| Energy contribution | -38.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687533 106 + 23771897 AAGCUCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGA-----------UC---ACC ((((.(((....)))))))....((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).)))))).((((..-----------.)---))) ( -39.90) >DroSec_CAF1 75322 106 + 1 AAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGA-----------UC---ACC (((((.........)))))....((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).)))))).((((..-----------.)---))) ( -40.80) >DroSim_CAF1 75801 106 + 1 AAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGA-----------UC---ACC (((((.........)))))....((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).)))))).((((..-----------.)---))) ( -40.80) >DroEre_CAF1 79683 106 + 1 AAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGA-----------UC---ACC (((((.........)))))....((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).)))))).((((..-----------.)---))) ( -40.80) >DroYak_CAF1 77933 117 + 1 AAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGAUCACGGGUGGAUC---ACC ..((((((.(((((.(.......((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).))))))).)))))..)))))).....---... ( -49.31) >DroAna_CAF1 72488 109 + 1 AAGCCCGUUUCCAUGGCUUGGUUUUUGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAACGAAGAA-----------CCUCCACC (((((.........)))))((((((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).))))).....)))-----------))...... ( -39.50) >consensus AAGCCCGUUUCCAUGGCUUGGUUUUCGGUUGCGCCUCGCUAGCCGGCCUCCUUUAUGCUGGGCUGGCUGCCUUGCCCACGCGUGCGCUACCGAAUGGUGGA___________UC___ACC (((((.........)))))....((((((.((((..(((.(((((((((..........)))))))))((...))....))).)))).)))))).......................... (-38.48 = -38.20 + -0.28)

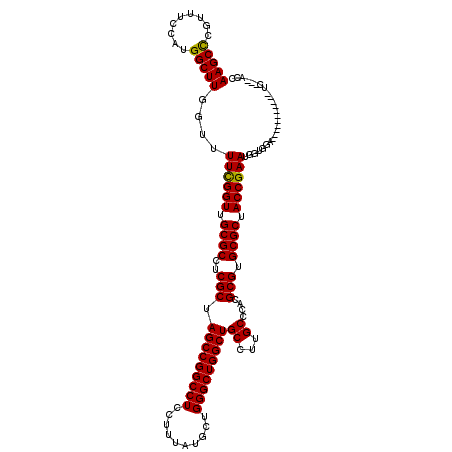
| Location | 8,687,533 – 8,687,639 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -39.87 |
| Energy contribution | -40.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8687533 106 - 23771897 GGU---GA-----------UCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGAGCUU (((---(.-----------..)))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....((((..((....)).)))) ( -40.72) >DroSec_CAF1 75322 106 - 1 GGU---GA-----------UCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUU (((---(.-----------..)))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....(((((.((....))))))) ( -45.22) >DroSim_CAF1 75801 106 - 1 GGU---GA-----------UCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUU (((---(.-----------..)))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....(((((.((....))))))) ( -45.22) >DroEre_CAF1 79683 106 - 1 GGU---GA-----------UCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUU (((---(.-----------..)))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....(((((.((....))))))) ( -45.22) >DroYak_CAF1 77933 117 - 1 GGU---GAUCCACCCGUGAUCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUU (((---(((((....).))).)))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....(((((.((....))))))) ( -48.42) >DroAna_CAF1 72488 109 - 1 GGUGGAGG-----------UUCUUCGUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCAAAAACCAAGCCAUGGAAACGGGCUU ((((((.(-----------.....).)))(((.((((.(((...(((..(((..((.............))..))).))).)))..)))).)))....)))(((((.((....))))))) ( -41.82) >consensus GGU___GA___________UCCACCAUUCGGUAGCGCACGCGUGGGCAAGGCAGCCAGCCCAGCAUAAAGGAGGCCGGCUAGCGAGGCGCAACCGAAAACCAAGCCAUGGAAACGGGCUU (((...((((.......)))).))).((((((.((((.(((...(((..(((..((.............))..))).))).)))..)))).))))))....(((((.((....))))))) (-39.87 = -40.37 + 0.50)

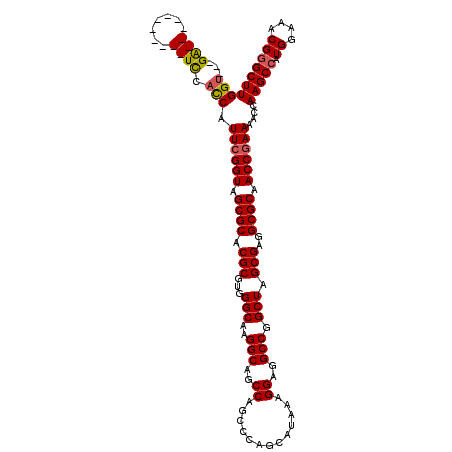

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:39 2006