| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
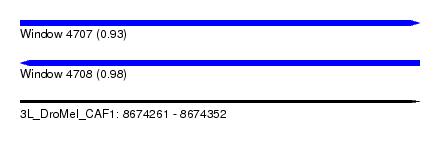
| Location | 8,674,261 – 8,674,352 |
| Length | 91 |
| Max. P | 0.982360 |

| Location | 8,674,261 – 8,674,352 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8674261 91 + 23771897 AGUUUUUUGUUAUUUUCUUAUUUU-----------------------CGUACAAAUGUACUGUUUGCCUGUGCAAAAUCUAUCUUCAUACGUACAUACGUACAUAUGUGUGUAC ........................-----------------------.((((....))))..(((((....)))))..............((((((((((....)))))))))) ( -16.80) >DroSec_CAF1 62356 110 + 1 AGUGUUUUGUUAUUUUCUUAUUUUCGCCCUGGGCAAACACACAUACUCGUACAAAUGUGCUGUUUGCCUGUGCAACAUCUAUGUACAUACGUAC----GAACGUAUGUGUGUAC .........................((.(.((((((((((((((..........))))).)))))))))).)).........((((((((((((----....)))))))))))) ( -34.50) >DroSim_CAF1 62752 110 + 1 AGUUUUUUGUUAUUUUCUUAUUUUCGCCCUGGGCAAACAUACAUACUCGUACAAAUGUGCUGUUUGCCUGUGCAACAUCUAUGUACAUACGUAC----GAACGUAUGUGUGUAC .........................((.(.((((((((((((((..........))))).)))))))))).)).........((((((((((((----....)))))))))))) ( -32.60) >consensus AGUUUUUUGUUAUUUUCUUAUUUUCGCCCUGGGCAAACA_ACAUACUCGUACAAAUGUGCUGUUUGCCUGUGCAACAUCUAUGUACAUACGUAC____GAACGUAUGUGUGUAC ................................................((((....))))...((((....)))).......((((((((((((........)))))))))))) (-14.82 = -15.60 + 0.78)

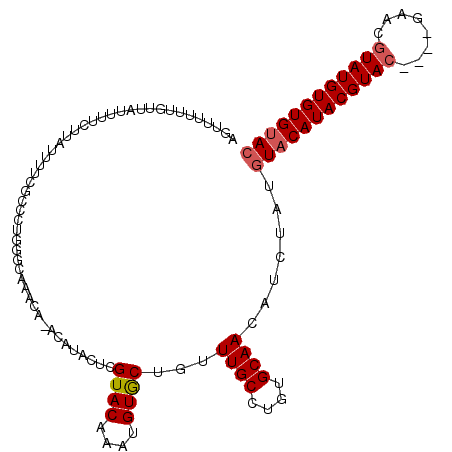

| Location | 8,674,261 – 8,674,352 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -15.37 |
| Energy contribution | -16.40 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8674261 91 - 23771897 GUACACACAUAUGUACGUAUGUACGUAUGAAGAUAGAUUUUGCACAGGCAAACAGUACAUUUGUACG-----------------------AAAAUAAGAAAAUAACAAAAAACU (((((..(((((((((....))))))))).........(((((....))))).........))))).-----------------------........................ ( -18.50) >DroSec_CAF1 62356 110 - 1 GUACACACAUACGUUC----GUACGUAUGUACAUAGAUGUUGCACAGGCAAACAGCACAUUUGUACGAGUAUGUGUGUUUGCCCAGGGCGAAAAUAAGAAAAUAACAAAACACU ..((((((((((..((----(((((.((((.......((((((....)).))))..)))).)))))))))))))))))(((((...)))))....................... ( -31.30) >DroSim_CAF1 62752 110 - 1 GUACACACAUACGUUC----GUACGUAUGUACAUAGAUGUUGCACAGGCAAACAGCACAUUUGUACGAGUAUGUAUGUUUGCCCAGGGCGAAAAUAAGAAAAUAACAAAAAACU ......((((((((..----..)))))))).........((((...(((((((((((.((((....)))).))).))))))))....))))....................... ( -27.70) >consensus GUACACACAUACGUUC____GUACGUAUGUACAUAGAUGUUGCACAGGCAAACAGCACAUUUGUACGAGUAUGU_UGUUUGCCCAGGGCGAAAAUAAGAAAAUAACAAAAAACU (((((.((((((((........)))))))).........((((....))))..........)))))...........(((((.....)))))...................... (-15.37 = -16.40 + 1.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:30 2006