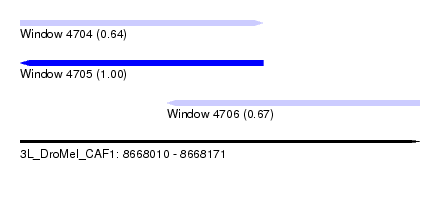
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,668,010 – 8,668,171 |
| Length | 161 |
| Max. P | 0.999969 |

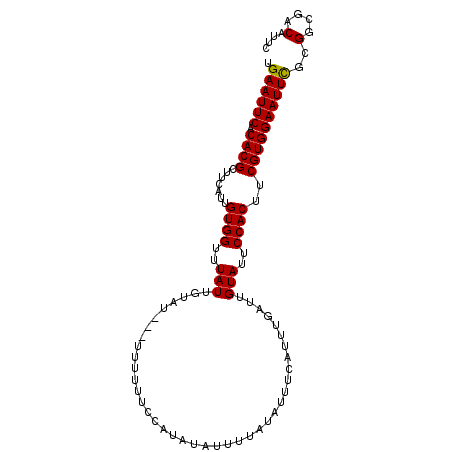
| Location | 8,668,010 – 8,668,108 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8668010 98 + 23771897 UGAAUUCACACGCUUCGUUGUGGUUUAUUGUAU---UUUUUUCCAUAUAAUUUAUAUAUCAUUUGAUUGUAUUCCACUUCGUGGAAUUUGCGGCGACAUUC .((((((.(.(((.(((..(((((..(((((((---........)))))))......))))).)))....(((((((...)))))))..)))).)).)))) ( -17.10) >DroSec_CAF1 55872 98 + 1 UGAAUUCACACGCUUCGUUGUGGUUUAUUGUAU---UUUUUUCCAUAUAUUUUAUAUUUCAUUUUAUUGUAUUCCACUUCGUGGAAUUCGCGGCGACAUUC .((((((.((((.......((((..(((.(((.---........(((.....))).........))).)))..))))..))))))))))..(....).... ( -17.07) >DroSim_CAF1 56227 98 + 1 UGAAUUCACACGCUUCAUUGUGGUUUAUUGUAU---UUUUUUCCAUAUAUUUUAUAUUUCAUUUGAUUGUAUUCCACUUCGUGGAAUUCGCGGCGACAUUC .((((((.(.(((.(((..((((..(((.((((---..........))))...)))..)))).)))..(.(((((((...))))))).))))).)).)))) ( -17.70) >DroEre_CAF1 57245 100 + 1 UGAAUUCACACGCU-CAUUGUGGUUUAUUGUACGUAUUUCUUCCAUCUAUUUUUUAUUCCAUUUGAUUGUAUUCCACUUCGUGGAAUUCGCGGCGACAUUC .((((((.(.((((-((..((((..((...........................))..)))).)))..(.(((((((...))))))).))))).)).)))) ( -17.03) >DroYak_CAF1 57698 97 + 1 UGAAUUCACACGCUUCAUUGUGGUUUAUUGU----AUUUUUUCCAUCUAUUUUUUAUUCCAUUCGAUUGUAUUCCACUUCGUGGAAUUCGCGGCGACAUUC .((((((.(.(((.((...((((..((....----...................))..))))..))..(.(((((((...))))))).))))).)).)))) ( -15.60) >consensus UGAAUUCACACGCUUCAUUGUGGUUUAUUGUAU___UUUUUUCCAUAUAUUUUAUAUUUCAUUUGAUUGUAUUCCACUUCGUGGAAUUCGCGGCGACAUUC .((((((.((((.......((((..(((........................................)))..))))..))))))))))..(....).... (-14.48 = -14.32 + -0.16)

| Location | 8,668,010 – 8,668,108 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -18.51 |
| Energy contribution | -18.51 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.03 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8668010 98 - 23771897 GAAUGUCGCCGCAAAUUCCACGAAGUGGAAUACAAUCAAAUGAUAUAUAAAUUAUAUGGAAAAAA---AUACAAUAAACCACAACGAAGCGUGUGAAUUCA ((((.(((((((..(((((((...)))))))....(((.(((((......))))).)))......---....................))).)))))))). ( -21.00) >DroSec_CAF1 55872 98 - 1 GAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUAAAAUGAAAUAUAAAAUAUAUGGAAAAAA---AUACAAUAAACCACAACGAAGCGUGUGAAUUCA ((((.((((((((.(((((((...))))))).).......................(((......---..........))).......))).)))))))). ( -19.09) >DroSim_CAF1 56227 98 - 1 GAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAAAUGAAAUAUAAAAUAUAUGGAAAAAA---AUACAAUAAACCACAAUGAAGCGUGUGAAUUCA ((((.((((((((.(((((((...))))))).).......................(((......---..........))).......))).)))))))). ( -19.09) >DroEre_CAF1 57245 100 - 1 GAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAAAUGGAAUAAAAAAUAGAUGGAAGAAAUACGUACAAUAAACCACAAUG-AGCGUGUGAAUUCA ((((.((((((((.(((((((...))))))).)..(((..(((............(((.........)))........)))...))-)))).)))))))). ( -20.05) >DroYak_CAF1 57698 97 - 1 GAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCGAAUGGAAUAAAAAAUAGAUGGAAAAAAU----ACAAUAAACCACAAUGAAGCGUGUGAAUUCA ((((.(((((((..(((((((((..((.....)).)))..))))))..........(((.......----........))).......))).)))))))). ( -19.56) >consensus GAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAAAUGAAAUAUAAAAUAUAUGGAAAAAA___AUACAAUAAACCACAAUGAAGCGUGUGAAUUCA ((((.(((((((..(((((((...))))))).........................(((...................))).......))).)))))))). (-18.51 = -18.51 + -0.00)

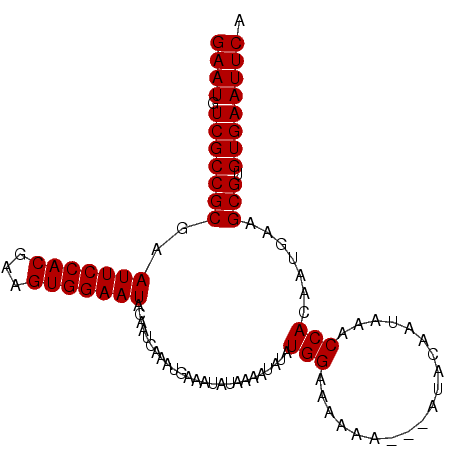
| Location | 8,668,069 – 8,668,171 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
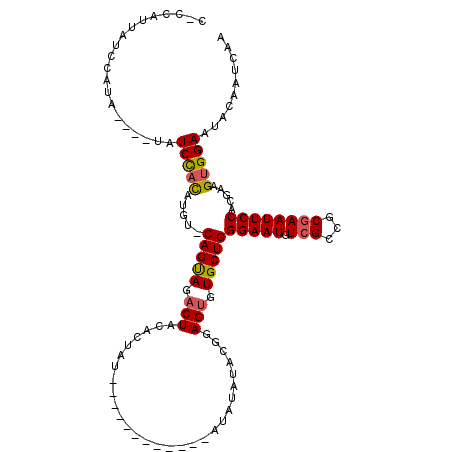
>3L_DroMel_CAF1 8668069 102 - 23771897 CCCCAUUAUCCAUA----UAUCCACAUGU-CAGUAGACUACACUAUAU----------AUAUAUACGAAGUGUGCUGGGAAUGUCGCCGCAAAUUCCACGAAGUGGAAUACAAUCAA ..............----..(((((....-(((((.(((.(..((((.----------...)))).).))).)))))(((((..........))))).....))))).......... ( -20.30) >DroSec_CAF1 55931 99 - 1 C-CCAUUAUCGAUA----UAUCUACAUGU-CAGUAGACUACACUAU------------AUAUAUACGGAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUAAA .-(((((.(((...----..(((((....-..))))).........------------........(((((.(((.(((.....).))))).))))).))))))))........... ( -22.80) >DroSim_CAF1 56286 92 - 1 C-CCAUUAUCCAUA----UAUCUACAUGU-CAGUAGACU-------------------AUAUAUACGGAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAA .-(((((.((.(((----(((((((....-..))))).)-------------------))))....(((((.(((.(((.....).))))).)))))..)))))))........... ( -22.20) >DroEre_CAF1 57306 114 - 1 C-CCAUUAUCCAUAUAUGUAUCCACAUGU-CAGUAGACUAAGCUACAUACAUACUCGUAUAUAUAC-GAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAA .-..............(((((((((...(-(.((((......))))...((((((((((....)))-)))))))...(((((.(((...))))))))..)).)))).)))))..... ( -32.30) >DroYak_CAF1 57756 104 - 1 C-CCAUUAUCCAUACAUAUAUCCACAUGU-CAGCAGACUACAGUA-----------GACUAUAUACGGAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCGA .-..................(((((....-(((((.(((...(((-----------.......)))..))).)))))(((((.(((...)))))))).....))))).......... ( -27.20) >DroAna_CAF1 53744 98 - 1 --CCAUUAUCCAUA----UAUCCAUAAAUCCAGCAGUCUAUAUAAU------------AGGUGC-UAGAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGCGGAAUACAAUCAA --............----...........((((((.(((((((...------------..))).-))))...))))))((.(((..((((............))))...))).)).. ( -20.50) >consensus C_CCAUUAUCCAUA____UAUCCACAUGU_CAGUAGACUACACUAU____________AUAUAUACGGAGUGUGCUGGGAAUGUCGCCGCGAAUUCCACGAAGUGGAAUACAAUCAA ....................(((((.....(((((.(((.............................))).)))))(((((.(((...)))))))).....))))).......... (-17.60 = -17.52 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:28 2006