
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,652,460 – 8,652,643 |
| Length | 183 |
| Max. P | 0.987322 |

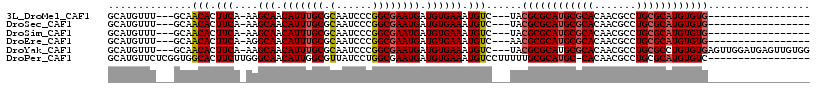
| Location | 8,652,460 – 8,652,553 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8652460 93 - 23771897 GCAUGUUU---GCAACACUUCA-AAGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC---UACGCGCAUGCGCACAACGCCUGCGCAUGUGUG----------------- (((....)---)).(((.(((.-..(((.(((((((.(......)))))))).)))))).))).---..((((((((((((.......))))))))))))----------------- ( -33.30) >DroSec_CAF1 40379 93 - 1 GCAUGUUU---GCAACACUUCA-AAGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC---UACGCGCAUGCGCACAACGCCUGCGCAUGUGUG----------------- (((....)---)).(((.(((.-..(((.(((((((.(......)))))))).)))))).))).---..((((((((((((.......))))))))))))----------------- ( -33.30) >DroSim_CAF1 40751 93 - 1 GCAUGUUU---GCAACACUUCA-AAGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC---UACGCGCAUGCGCACAACGCCUGCGCAUGUGUG----------------- (((....)---)).(((.(((.-..(((.(((((((.(......)))))))).)))))).))).---..((((((((((((.......))))))))))))----------------- ( -33.30) >DroEre_CAF1 41827 93 - 1 GCAUGUUU---GCAACACUUCA-AGGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC---AACGCGCAUGCGCACAACGCCUGCGCAUGUGUG----------------- (((....)---)).(((.(((.-..(((.(((((((.(......)))))))).)))))).))).---..((((((((((((.......))))))))))))----------------- ( -33.30) >DroYak_CAF1 41599 110 - 1 GCAUGUUU---GCAACACUUCA-AAGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC---UACGCGCAUGCGCACAACGCCUGCGCCUGUGUGAGUUGGAUGAGUUGUGG ......(.---.((((.(((((-(.(((.(((((((.(......)))))))).)))........---..((((((.(((((.......))))).))))))..))))).).))))..) ( -34.20) >DroPer_CAF1 43617 99 - 1 GCAUGUUCUCGGUGGCACUUCUUGGGCAACAUUGGCGUUAUCCUGGCGAAUGAUGUGAAAUGUCCUUUUUGCGCAUGC-CACAACGCCUGCGCAUGUGUC----------------- .............(((((....((.(((.....((((((....((((..(((.((..(((......)))..)))))))-)).))))))))).)).)))))----------------- ( -28.20) >consensus GCAUGUUU___GCAACACUUCA_AAGCAACAUUUGCGCAAUCCCGGCGAAUGAUGUGAAAUGUC___UACGCGCAUGCGCACAACGCCUGCGCAUGUGUG_________________ ..............(((.(((....(((.(((((((.(......)))))))).)))))).)))......((((((((((((.......))))))))))))................. (-27.39 = -27.78 + 0.39)


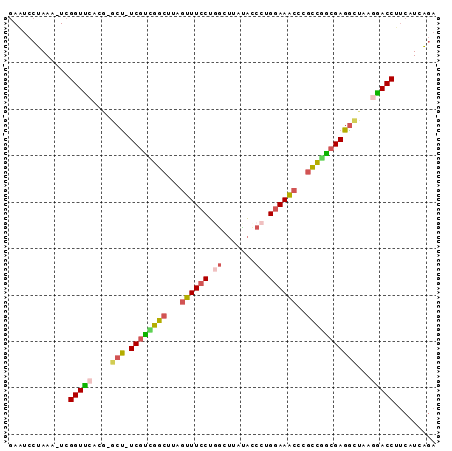
| Location | 8,652,553 – 8,652,643 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.46 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8652553 90 + 23771897 GAAUCCUAAACUCGGUUCACA-ACU-UCGUUGGCUUAGUUUCCUGGCUUAUAUCCUGGAAACCCGUCGACGAGGCUAAGGACCUUCAUCAGA .............(((((...-.((-((((((((...((((((.((.......)).))))))..))))))))))....)))))......... ( -28.80) >DroSec_CAF1 40472 89 + 1 GAAUCCUAAA-UCGGUUCACG-GCU-UCGUUGGCUUAGUUUCCUGGCUUAUACCCUGGAAACCCGCUGGCGAGGCUAAAAACCUUCAUCAGA ..........-..((((...(-(((-((((..((...((((((.((.......)).))))))..))..))))))))...))))......... ( -32.00) >DroSim_CAF1 40844 89 + 1 GAAUCCUAAA-UCGGUUCACG-GCUUUCGUGGGCUUAGUUUCCUGGCUUAUACCCUG-AAACACGCUGGCGAGGCUAAGAACCUUCAUCAGA ..........-..(((((..(-((((.(((.(((...(((((..((......))..)-))))..))).))))))))..)))))......... ( -27.60) >DroEre_CAF1 41920 89 + 1 GAGUCCUAAC-UCGGUCCACG-GCU-UCGUCGGCUUAGUUUCCUGGCUUACAUCCGGGAAACCCGCCGGCGAGGCUACGGACCUCCAUCAGA ((((....))-))(((((..(-(((-((((((((...(((((((((.......)))))))))..))))))))))))..)))))......... ( -48.20) >DroYak_CAF1 41709 90 + 1 GAAUCCUAAC-UCGGUCCACGGGCU-UCGUCGGCUCAGUUUCCUCGCUUAUAUCCUGGAAACCAGCCGGCGAGACUAAGGACCUUCAUCAGA ..........-..(((((...((..-(((((((((..((((((.............)))))).)))))))))..))..)))))......... ( -34.42) >DroAna_CAF1 38597 79 + 1 UAA---GAAA-UCGGUUGAGA-ACC-UCCCAGG-----GUUCCACAUCUAUUCCCAGGAAUAAUCCCUGCGAGGUCA-GGACC-UCAUCUCA ...---....-..((((..((-.((-((.((((-----(.........(((((....)))))..))))).)))))).-.))))-........ ( -23.20) >consensus GAAUCCUAAA_UCGGUUCACG_GCU_UCGUCGGCUUAGUUUCCUGGCUUAUACCCUGGAAACCCGCCGGCGAGGCUAAGGACCUUCAUCAGA .............(((((....(((.((((((((...((((((.((.......)).))))))..)))))))))))...)))))......... (-24.27 = -24.22 + -0.05)


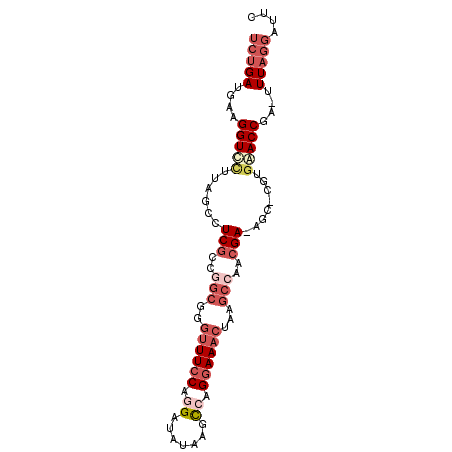
| Location | 8,652,553 – 8,652,643 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -14.71 |
| Energy contribution | -16.91 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8652553 90 - 23771897 UCUGAUGAAGGUCCUUAGCCUCGUCGACGGGUUUCCAGGAUAUAAGCCAGGAAACUAAGCCAACGA-AGU-UGUGAACCGAGUUUAGGAUUC (((((....(((.(.((((.((((.(...(((((((.((.......)).)))))))....).))))-.))-)).).)))....))))).... ( -24.30) >DroSec_CAF1 40472 89 - 1 UCUGAUGAAGGUUUUUAGCCUCGCCAGCGGGUUUCCAGGGUAUAAGCCAGGAAACUAAGCCAACGA-AGC-CGUGAACCGA-UUUAGGAUUC (((((....(((((...((.(((...((.(((((((..(((....))).)))))))..))...)))-.))-...)))))..-.))))).... ( -27.20) >DroSim_CAF1 40844 89 - 1 UCUGAUGAAGGUUCUUAGCCUCGCCAGCGUGUUU-CAGGGUAUAAGCCAGGAAACUAAGCCCACGAAAGC-CGUGAACCGA-UUUAGGAUUC (((((....(((((...((.(((...((..((((-(..(((....)))..)))))...))...)))..))-...)))))..-.))))).... ( -25.90) >DroEre_CAF1 41920 89 - 1 UCUGAUGGAGGUCCGUAGCCUCGCCGGCGGGUUUCCCGGAUGUAAGCCAGGAAACUAAGCCGACGA-AGC-CGUGGACCGA-GUUAGGACUC ((((((...((((((..((.(((.((((.(((((((.((.......)).)))))))..)))).)))-.))-..))))))..-)))))).... ( -43.30) >DroYak_CAF1 41709 90 - 1 UCUGAUGAAGGUCCUUAGUCUCGCCGGCUGGUUUCCAGGAUAUAAGCGAGGAAACUGAGCCGACGA-AGCCCGUGGACCGA-GUUAGGAUUC ((((((...(((((...(..(((.((((((((((((.(........)..))))))).))))).)))-..)....)))))..-)))))).... ( -36.60) >DroAna_CAF1 38597 79 - 1 UGAGAUGA-GGUCC-UGACCUCGCAGGGAUUAUUCCUGGGAAUAGAUGUGGAAC-----CCUGGGA-GGU-UCUCAACCGA-UUUC---UUA .(((((..-(((..-.((((((.(((((....((((.............)))))-----)))).))-)))-)....))).)-))))---... ( -27.92) >consensus UCUGAUGAAGGUCCUUAGCCUCGCCGGCGGGUUUCCAGGAUAUAAGCCAGGAAACUAAGCCAACGA_AGC_CGUGAACCGA_UUUAGGAUUC (((((....(((((......(((..(((..((((((.((.......)).))))))...)))..)))........)))))....))))).... (-14.71 = -16.91 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:16 2006