
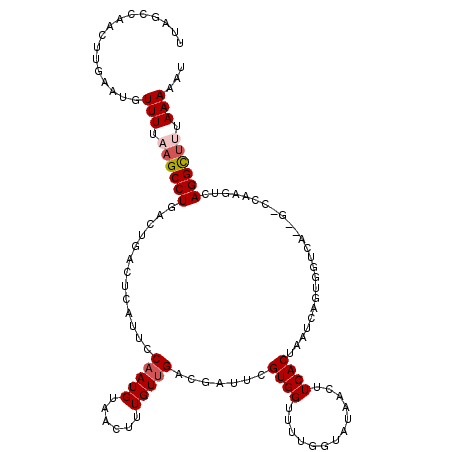
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,648,268 – 8,648,388 |
| Length | 120 |
| Max. P | 0.955829 |

| Location | 8,648,268 – 8,648,388 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8648268 120 + 23771897 UUAGCCAACUUGAAUGUUUAAAGCCUGACUGACUCAUUCCAAUGUAACUUUGUUGACGAUUCGUGGUUUUGGUAUAACUUCACUAAUCAGUGGUCAGAGUCCAAGUCAGGCUUUAAAAAU ................(((((((((((((((((((....(((.......))).((((.(((..((((...((.....))..))))...))).)))))))))..))))))))))))))... ( -35.60) >DroSec_CAF1 36297 101 + 1 UUAGCCAACUUGAAUGUUUUAAGCCUGACUGACUCAUUCCAAUGUAACUUUGUUGACGAGUCGUGGUUUUGUUAUAACUUCACUAAUC-------------------AGGCUUUAAAAAU ................((((((((((((..(((((....(((((......)))))..)))))((((.............))))...))-------------------)))).)))))).. ( -21.92) >DroSim_CAF1 33704 101 + 1 UUAGCCAACUUGAAUGUUUUAAGCCUGACUGACUCAUUCCAAUGUAACUUUGUUGACGAGUCGUGGUUUUGUUAUAACUUCACUAAUC-------------------AGGCUUUAAAAAU ................((((((((((((..(((((....(((((......)))))..)))))((((.............))))...))-------------------)))).)))))).. ( -21.92) >DroEre_CAF1 37630 120 + 1 UUAGGCAACUUGAAUGUUUUUAGCCUAACUGACUCAUUCCAAUGUAACUUUGUUGACAAUUCGUGGUUUUGGUAUAACAUCACUAAUAAGUGGUCACGGCCCAAGUCAGGUUUUAAAAAC (((((((((......)))....))))))((((((.....(((((......))))).....((((((((.......)))((((((....)))))))))))....))))))........... ( -24.80) >DroYak_CAF1 37385 108 + 1 UAA------------GUUUUUUGCCUGACUGACUCAUUCCCAUGUAACUUUGUUGACCAUUUGUGGUUUCGGUAUAACUUCACUAAUAAGUGGUCAGAGUCCAAGUCAGGUUUUAAAAUU ..(------------(((((..(((((((((((((.....((........)).((((((((((((((...((.....))..))).))))))))))))))))..))))))))...)))))) ( -34.60) >consensus UUAGCCAACUUGAAUGUUUUAAGCCUGACUGACUCAUUCCAAUGUAACUUUGUUGACGAUUCGUGGUUUUGGUAUAACUUCACUAAUCAGUGGUCA__G_CCAAGUCAGGCUUUAAAAAU ................(((.((((((.............(((((......))))).......((((.............))))........................)))))).)))... ( -8.14 = -8.70 + 0.56)


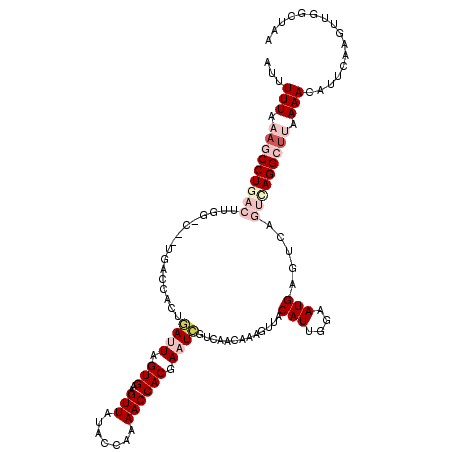
| Location | 8,648,268 – 8,648,388 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -8.36 |
| Energy contribution | -10.84 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8648268 120 - 23771897 AUUUUUAAAGCCUGACUUGGACUCUGACCACUGAUUAGUGAAGUUAUACCAAAACCACGAAUCGUCAACAAAGUUACAUUGGAAUGAGUCAGUCAGGCUUUAAACAUUCAAGUUGGCUAA ...((((((((((((((..(((((...(((..((((.(((..(((.......)))))).))))((.(((...)))))..)))...)))))))))))))))))))................ ( -33.60) >DroSec_CAF1 36297 101 - 1 AUUUUUAAAGCCU-------------------GAUUAGUGAAGUUAUAACAAAACCACGACUCGUCAACAAAGUUACAUUGGAAUGAGUCAGUCAGGCUUAAAACAUUCAAGUUGGCUAA .......((((((-------------------((((.(((..(((.......))))))(((((((...(((.......)))..))))))))))))))))).................... ( -25.20) >DroSim_CAF1 33704 101 - 1 AUUUUUAAAGCCU-------------------GAUUAGUGAAGUUAUAACAAAACCACGACUCGUCAACAAAGUUACAUUGGAAUGAGUCAGUCAGGCUUAAAACAUUCAAGUUGGCUAA .......((((((-------------------((((.(((..(((.......))))))(((((((...(((.......)))..))))))))))))))))).................... ( -25.20) >DroEre_CAF1 37630 120 - 1 GUUUUUAAAACCUGACUUGGGCCGUGACCACUUAUUAGUGAUGUUAUACCAAAACCACGAAUUGUCAACAAAGUUACAUUGGAAUGAGUCAGUUAGGCUAAAAACAUUCAAGUUGCCUAA ...........((((((((..(((((((((((....)))).....................(((....))).)))))...))..))))))))((((((....(((......))))))))) ( -25.20) >DroYak_CAF1 37385 108 - 1 AAUUUUAAAACCUGACUUGGACUCUGACCACUUAUUAGUGAAGUUAUACCGAAACCACAAAUGGUCAACAAAGUUACAUGGGAAUGAGUCAGUCAGGCAAAAAAC------------UUA ..........(((((((..(((((((((((((....))))..))))..((...((((....))))...((........))))...))))))))))))........------------... ( -23.60) >consensus AUUUUUAAAGCCUGACUUGG_C__UGACCACUGAUUAGUGAAGUUAUACCAAAACCACGAAUCGUCAACAAAGUUACAUUGGAAUGAGUCAGUCAGGCUUAAAACAUUCAAGUUGGCUAA ...(((.(((((((((................((((.(((..(((.......)))))).)))).............(((....))).....))))))))).)))................ ( -8.36 = -10.84 + 2.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:10 2006