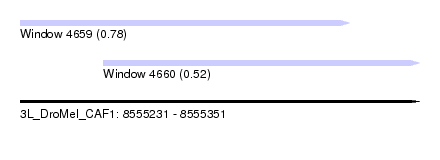
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,555,231 – 8,555,351 |
| Length | 120 |
| Max. P | 0.783108 |

| Location | 8,555,231 – 8,555,330 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8555231 99 + 23771897 CUAGACAUCAGUCAAUUAUGAAGUCAACUGCUAUUCUUAGCAGGCACC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCUACCCUUCAAUU ...(((....))).....(((((....((((((....)))))).....-(((.((((((((...((....)).....))))))..)-).))).)))))... ( -19.50) >DroSec_CAF1 6522 91 + 1 CUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCACC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAU---------ACCCUUCAAUU ...(((....))).....(((((....((((((....)))))).....-...........(((((((.........)))))---------)).)))))... ( -19.80) >DroSim_CAF1 6975 99 + 1 CUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCAGC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCUACCCUUCAAUU ...(((....))).....(((((......((((....))))(((.(((-.(((((((..........))))))).........)))-)))...)))))... ( -19.70) >DroEre_CAF1 6842 99 + 1 CUAGACAUCAGUCAACUAUGAAGUCAGCUGCUAUUCUUAGCAGGCACC-GUAUGAAUACUAUAUUGGAAUCAUAAAUCAAUAAGCU-CCUGCCCUUCAAUU ...(((....))).....(((((.(((((((((....))))))((...-(((....)))..((((((.........)))))).)).-.)))..)))))... ( -19.10) >DroYak_CAF1 6762 99 + 1 CUAGACAUCGGUCAACUAUGAAGUCAACUGCUAUUUUUAGCAGGCAGC-GUAUGAUUACUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCAACCCUUCAAUU ...(((....))).....(((((....((((((....))))))(.(((-.(((((((.(......).))))))).........)))-.)....)))))... ( -21.50) >DroPer_CAF1 8688 97 + 1 C----CAUCAUUGAGUGGUGAAGUCAUUUGCUAUUCCCAGCAGUCACAUUUAUGAUUAUAAUAUUACAAUCAUAAAUCUAUAGGCUGUCGAGACUUCAAUU (----(((......))))(((((((....(((......)))((((..((((((((((.((....)).)))))))))).....)))).....)))))))... ( -22.50) >consensus CUAGACAUCAGUCAACUAUGAAGUCAACUGCUAUUCUUAGCAGGCACC_GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU_CCUACCCUUCAAUU ...(((....))).....(((((....((((((....)))))).......(((((((.(......).)))))))...................)))))... (-15.72 = -16.05 + 0.33)



| Location | 8,555,256 – 8,555,351 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -18.89 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8555256 95 + 23771897 AACUGCUAUUCUUAGCAGGCACC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCUACCCUUCAAUUUGAUAUUGCAGCUA---------------CUUUGGU ..((((((....))))))..(((-((((((((..........)))))))........((((-.(......((.....))....).)))).---------------...)))) ( -16.90) >DroSec_CAF1 6547 87 + 1 AACUGCUAUUCUUAGCAGGCACC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAU---------ACCCUUCAAUUUGAUAUUGCAGCUA---------------CUUUGGU ..((((((....))))))..(((-(..........(((((((.........)))))---------)).((.((((.....)))).))...---------------...)))) ( -16.40) >DroSim_CAF1 7000 95 + 1 AACUGCUAUUCUUAGCAGGCAGC-GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCUACCCUUCAAUUUGAUAUUGCAGCUA---------------CUUUGGU ..((((((....))))))((.((-((((((((..........))))))).(((((.(((..-......)))....)))))..))).))..---------------....... ( -16.80) >DroEre_CAF1 6867 95 + 1 AGCUGCUAUUCUUAGCAGGCACC-GUAUGAAUACUAUAUUGGAAUCAUAAAUCAAUAAGCU-CCUGCCCUUCAAUUUGAUAUUGCAGCUA---------------CUUUGGU ((((((........(((((....-(((....)))..((((((.........))))))....-)))))...((.....))....)))))).---------------....... ( -17.20) >DroYak_CAF1 6787 95 + 1 AACUGCUAUUUUUAGCAGGCAGC-GUAUGAUUACUGUAUUGAAAUCAUAAAUCAAUAAGCU-CCAACCCUUCAAUUUGAUAUUGCAGCUA---------------CUUUGGC ..((((((....))))))...((-.(((((((.(......).)))))))...(((..((((-.(((....((.....))..))).)))).---------------..))))) ( -19.70) >DroPer_CAF1 8709 112 + 1 AUUUGCUAUUCCCAGCAGUCACAUUUAUGAUUAUAAUAUUACAAUCAUAAAUCUAUAGGCUGUCGAGACUUCAAUUUCAUACUGCAGCUAAUCGAAGUCUCAAAGCUUCUGA ....(((.....((((......((((((((((.((....)).))))))))))......))))..((((((((.(((.............))).))))))))..)))...... ( -26.32) >consensus AACUGCUAUUCUUAGCAGGCACC_GUAUGAUUAUUGUAUUGAAAUCAUAAAUCAAUAAGCU_CCUACCCUUCAAUUUGAUAUUGCAGCUA_______________CUUUGGU ..((((((....))))))(((....(((((((.(......).))))))).(((((....................)))))..)))........................... (-12.23 = -12.57 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:42 2006