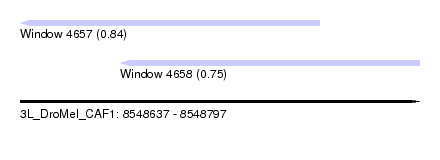
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,548,637 – 8,548,797 |
| Length | 160 |
| Max. P | 0.843309 |

| Location | 8,548,637 – 8,548,757 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -45.28 |
| Consensus MFE | -31.27 |
| Energy contribution | -30.25 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
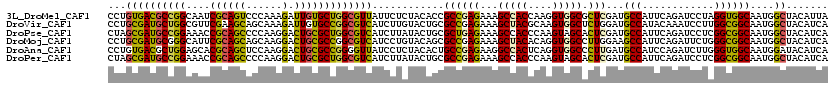
>3L_DroMel_CAF1 8548637 120 - 23771897 CCUGUGACGCCGGCAAUCGCAGUCCCAAAGAUUGUGCUGGCGUUAUUCUCUACACCGCCGAGAAAGCCACCAAGGUGGCGCUCGAUGCCAUUCAGAUCCUAGGUGGCAAUGGCUACAUUA ...(((((((((((....((((((.....)))))))))))))))))..........((((((...(((((....))))).)))..((((((..((...))..))))))..)))....... ( -50.90) >DroVir_CAF1 117 120 - 1 CCUGCGAUGCUGGCGUUCGAAGCAGCAAAGAUUGUGCCGGCGUCAUCUUGUACUGCGCCGAGAAAGCUACGCAAGUGGCUCUGGAUGCCAUACAAAUCCUUGGCGGCAAUGGCUACAUCA ..((((((((((((((.....)).(((.....))))))))))))...........(((((((..((((((....)))))).(((...)))........))))))))))............ ( -42.60) >DroPse_CAF1 117 120 - 1 CUAGCGAUGCCGGAAACCGCAGCCCCAAGGACUGCGCUGGCGUCAUCUUAUACUGCGCUGAGAAAGCCACCCAAGUAGCACUCGAUGCCAUUCAGAUCCUCGGCGGCAAUGGCUACAUCA .((((..(((.(....).)))(((((.(((((((...(((((((.(((((........)))))..((.((....)).))....)))))))..))).)))).)).)))....))))..... ( -42.50) >DroMoj_CAF1 117 120 - 1 CCUGCGAUGCGGGCAUUCGCAGCAGCAAGGACUGCGCCGGCGUCAUCCUGUACAGCGCCGAGAAAGCUACACAGGUGGCCUUGGAAGCCAUUCAGAUUCUGGGCGGCAAUGGCUACAUCA .....((((..(.((((((((((......).)))))((((.((((.(((((..(((.........)))..))))))))).))))..(((.(((((...))))).))))))).)..)))). ( -42.50) >DroAna_CAF1 117 120 - 1 CCUGUGACGCUGGAGCACGCAGCUCCAAGGACUGCGCCGGGGUUAUCCUCUACACUGCCGAGAAGGCCACUCAGGUGGCCCUUGAUGCCAUCCAGAUCUUGGGUGGCAAUGGAUACAUCA ((((.(.(((((((((.....))))))......))))))))..(((((..........((((..((((((....)))))))))).((((((((((...))))))))))..)))))..... ( -50.30) >DroPer_CAF1 1801 120 - 1 CUAGCGAUGCCGGAAACCGCAGCCCCAAGGACUGCGCUGGCGUCAUCUUAUACUGCGCCGAGAAAGCCACCCAAGUAGCACUCGAUGCCAUUCAGAUCCUCGGCGGCAAUGGCUACAUCA .((((((((((((....(((((((....)).))))))))))))).........(((((((((...((.((....)).))....((......)).....)))))).)))...))))..... ( -42.90) >consensus CCUGCGAUGCCGGAAAUCGCAGCCCCAAGGACUGCGCCGGCGUCAUCCUAUACAGCGCCGAGAAAGCCACCCAAGUGGCACUCGAUGCCAUUCAGAUCCUCGGCGGCAAUGGCUACAUCA ...((((((((((....((((((......).)))))))))))))............((((((...(((((....))))).)))...(((............))))))....))....... (-31.27 = -30.25 + -1.02)

| Location | 8,548,677 – 8,548,797 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -46.72 |
| Consensus MFE | -31.16 |
| Energy contribution | -30.05 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8548677 120 - 23771897 CUGAGUGCGUGCAGGAGCUAUUUGUACACGGUGGCCAGAUCCUGUGACGCCGGCAAUCGCAGUCCCAAAGAUUGUGCUGGCGUUAUUCUCUACACCGCCGAGAAAGCCACCAAGGUGGCG ....(((((.((....))....))))).((((((..(((....(((((((((((....((((((.....)))))))))))))))))..)))...)))))).....(((((....))))). ( -50.40) >DroVir_CAF1 157 120 - 1 CUGAGCGCGUGCCGGAGCUACUUGUAUGCUGUAGCAAGAUCCUGCGAUGCUGGCGUUCGAAGCAGCAAAGAUUGUGCCGGCGUCAUCUUGUACUGCGCCGAGAAAGCUACGCAAGUGGCU ((..((((((((.((((((((.........)))))..........(((((((((((.....)).(((.....)))))))))))).))).)))).))))..))..((((((....)))))) ( -48.70) >DroPse_CAF1 157 120 - 1 CUGAGCGCCUGUCGCAGCUAUCUCUAUACUGUGGCCAGAUCUAGCGAUGCCGGAAACCGCAGCCCCAAGGACUGCGCUGGCGUCAUCUUAUACUGCGCUGAGAAAGCCACCCAAGUAGCA ((.((((((((((((((.(((....))))))))).))).......((((((((....(((((((....)).)))))))))))))..........))))).))...((.((....)).)). ( -44.10) >DroMoj_CAF1 157 120 - 1 CUGGGCGCCUGCCGCAGCUAUUUGUAUUCAGUGGCCCGAGCCUGCGAUGCGGGCAUUCGCAGCAGCAAGGACUGCGCCGGCGUCAUCCUGUACAGCGCCGAGAAAGCUACACAGGUGGCC ((.(((((..((....))....(((((...((((((((.((((((...))))))....((.((((......))))))))).)))))...)))))))))).))...(((((....))))). ( -49.10) >DroAna_CAF1 157 120 - 1 CUGAGUGCCUGCCGGAGCUACCUAUACACCGUGGCCAGGUCCUGUGACGCUGGAGCACGCAGCUCCAAGGACUGCGCCGGGGUUAUCCUCUACACUGCCGAGAAGGCCACUCAGGUGGCC ((((((((((.((((((..........(((.(((((((.((((.......((((((.....))))))))))))).)))).)))...........)).))).).))).)))))))...... ( -47.71) >DroPer_CAF1 1841 120 - 1 CUGAGCGCCUGUCGCAGCUAUCUCUAUACUGUGGCCAGAUCUAGCGAUGCCGGAAACCGCAGCCCCAAGGACUGCGCUGGCGUCAUCUUAUACUGCGCCGAGAAAGCCACCCAAGUAGCA ((..(((((((((((((.(((....))))))))).))).......((((((((....(((((((....)).)))))))))))))..........))))..))...((.((....)).)). ( -40.30) >consensus CUGAGCGCCUGCCGCAGCUAUCUGUAUACUGUGGCCAGAUCCUGCGAUGCCGGAAAUCGCAGCCCCAAGGACUGCGCCGGCGUCAUCCUAUACAGCGCCGAGAAAGCCACCCAAGUGGCA ((..((((........(((((.........)))))..........((((((((....((((((......).)))))))))))))..........))))..))...(((((....))))). (-31.16 = -30.05 + -1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:40 2006