
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,488,847 – 8,489,003 |
| Length | 156 |
| Max. P | 0.996797 |

| Location | 8,488,847 – 8,488,939 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -15.58 |
| Energy contribution | -17.82 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
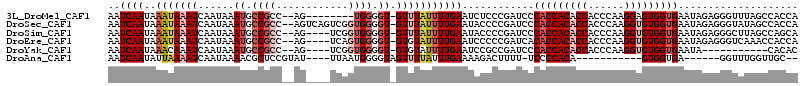
>3L_DroMel_CAF1 8488847 92 - 23771897 ACACCACCCAAGGAGUGGUGAAUAGAGGGUUUAGCCACCAUGCUCUUGUGACAGCAUAUUUCUCACACGAAGCAACGGUCAUACUUAUUCGC------------------------ .(((((((....).))))))....(((((.....))(((.(((((.(((((...........))))).).))))..)))....)))......------------------------ ( -22.90) >DroSec_CAF1 7275 92 - 1 ACACCACCCAAGGUGUGGUGAAUAGAGGGUAUAGCCACCAAGCUCUUGUGCCAGUAUAUCUCUCACACAGAGCAACGGUCAUACUCAUUUGC------------------------ .(((((((....).))))))......((((((..((.....(((((((((..((.....))..)))).)))))...))..))))))......------------------------ ( -31.00) >DroSim_CAF1 7241 92 - 1 ACACCACCCAAGGUGUGGUGAAUAGAGGGCUUAGCCAGCAUGCUCUUGUGCCAGUAUAUUUCUCACACGGAGCAACGGUCAUACUCAUUCGC------------------------ (((((......))))).((((((.((((((...)))....((((((.(((..((.......))..))))))))).........)))))))))------------------------ ( -27.50) >DroEre_CAF1 8297 116 - 1 ACACCACCCAAGGUGUGGUGAAUAGAGGGUCAAACCACCAUGCCCUUGUGCCAGUAUAUUUAUCAGAUGGAGCAACGGUCAUACUCAUUGACUCGUUUCCAUUGUGGUUGUAAAAA (((((((...(((((((.((....(((((.((........)))))))....)).)))))))....((((((..((((((((.......)))).)))))))))))))).)))..... ( -33.90) >DroYak_CAF1 7297 81 - 1 ACACCACCCAAGGUGUGGUGAAUA-----------CACACUGCCCUUGUGCCAGUAUAUUUAACAGUUGGAGCAACGGUCAUACUCAUUGGU------------------------ ........(((((.(..(((....-----------..)))..)))))).((((((.......((.((((...)))).)).......))))))------------------------ ( -19.84) >consensus ACACCACCCAAGGUGUGGUGAAUAGAGGGUUUAGCCACCAUGCUCUUGUGCCAGUAUAUUUCUCACACGGAGCAACGGUCAUACUCAUUCGC________________________ .(((((((....).))))))......((((...(((....((((((((((..((.....))..)))).))))))..)))...)))).............................. (-15.58 = -17.82 + 2.24)


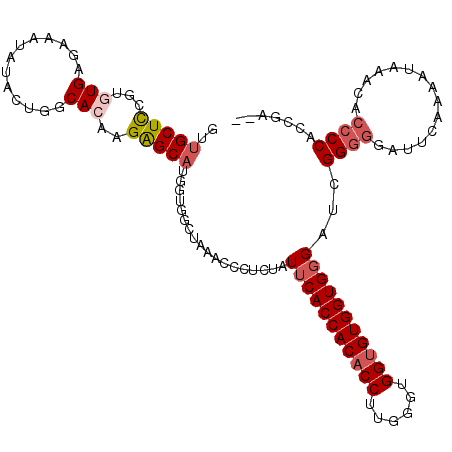
| Location | 8,488,863 – 8,488,972 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -40.87 |
| Consensus MFE | -26.02 |
| Energy contribution | -27.02 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8488863 109 + 23771897 GUUGCUUCGUGUGAGAAAUAUGCUGUCACAAGAGCAUGGUGGCUAAACCCUCUAUUCACCACUCCUUGGGUGGUGUGGUGGGAUCGGGAGAUUCAAAAUAAACACCCCA------ ..(((((..(((((.(.......).))))).))))).((((..((..(((.(((..(((((((.....)))))))))).)))(((....)))......))..))))...------ ( -35.60) >DroSec_CAF1 7291 115 + 1 GUUGCUCUGUGUGAGAGAUAUACUGGCACAAGAGCUUGGUGGCUAUACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGUAUUCAAAAUAAACACCCCACCGACU ...(((((((((.((.......)).)))).)))))(((((((..(((((((...(((((((((((......)))))))))))...)))))))..............))))))).. ( -49.69) >DroSim_CAF1 7257 113 + 1 GUUGCUCCGUGUGAGAAAUAUACUGGCACAAGAGCAUGCUGGCUAAGCCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGUAUUCAAAAUAAACACCCCACCGA-- ..(((((.((((.((.......)).))))..))))).(((.....)))......(((((((((((......)))))))))))...(((((.............))))).....-- ( -40.92) >DroEre_CAF1 8337 113 + 1 GUUGCUCCAUCUGAUAAAUAUACUGGCACAAGGGCAUGGUGGUUUGACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGUGAUCGGGGGAUUCAAAAUACACACCCCACUGA-- .(((..(((..............)))..)))(((....(((.((((((((((.((((((((((((......))))))))).))).)))))..)))))...)))..))).....-- ( -39.34) >DroYak_CAF1 7313 102 + 1 GUUGCUCCAACUGUUAAAUAUACUGGCACAAGGGCAGUGUG-----------UAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGCGGAUUCAAAAUACACACCCCACCGA-- ..(((((....(((((.......)))))...)))))(((((-----------(((((((((((((......)))))))))(((((....)))))..)))))))))........-- ( -38.80) >consensus GUUGCUCCGUGUGAGAAAUAUACUGGCACAAGAGCAUGGUGGCUAAACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGGAUUCAAAAUAAACACCCCACCGA__ ..(((((...(((.............)))..)))))..................(((((((((((......)))))))))))...((((...............))))....... (-26.02 = -27.02 + 1.00)


| Location | 8,488,863 – 8,488,972 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -26.06 |
| Energy contribution | -27.78 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8488863 109 - 23771897 ------UGGGGUGUUUAUUUUGAAUCUCCCGAUCCCACCACACCACCCAAGGAGUGGUGAAUAGAGGGUUUAGCCACCAUGCUCUUGUGACAGCAUAUUUCUCACACGAAGCAAC ------.(((..((((.....))))..))).....((((((.((......)).))))))....(..((.....))..).(((((.(((((...........))))).).)))).. ( -29.90) >DroSec_CAF1 7291 115 - 1 AGUCGGUGGGGUGUUUAUUUUGAAUACCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGUAUAGCCACCAAGCUCUUGUGCCAGUAUAUCUCUCACACAGAGCAAC ....((((((((((((.....))))))))).(((((((((((((......)))))))))......))))...))).....(((((((((..((.....))..)))).)))))... ( -46.50) >DroSim_CAF1 7257 113 - 1 --UCGGUGGGGUGUUUAUUUUGAAUACCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGCUUAGCCAGCAUGCUCUUGUGCCAGUAUAUUUCUCACACGGAGCAAC --..((((((((((((.....))))))))))..))(((((((((......)))))))))....(..(((...)))..).((((((.(((..((.......))..))))))))).. ( -45.10) >DroEre_CAF1 8337 113 - 1 --UCAGUGGGGUGUGUAUUUUGAAUCCCCCGAUCACACCACACCACCCAAGGUGUGGUGAAUAGAGGGUCAAACCACCAUGCCCUUGUGCCAGUAUAUUUAUCAGAUGGAGCAAC --.....((((((((...(((((..(((.(.....(((((((((......)))))))))....).))))))))....))))))))(((.(((..............))).))).. ( -37.54) >DroYak_CAF1 7313 102 - 1 --UCGGUGGGGUGUGUAUUUUGAAUCCGCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUA-----------CACACUGCCCUUGUGCCAGUAUAUUUAACAGUUGGAGCAAC --..((.(.((((((((((....(((....)))..(((((((((......)))))))))))))-----------)))))).)))((((.((((............)))).)))). ( -36.80) >consensus __UCGGUGGGGUGUUUAUUUUGAAUCCCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGUUUAGCCACCAUGCUCUUGUGCCAGUAUAUUUCUCACACGGAGCAAC ......(((((.((((.....)))).)))))....(((((((((......)))))))))....................((((((((((..((.....))..)))).)))))).. (-26.06 = -27.78 + 1.72)



| Location | 8,488,899 – 8,489,003 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -17.88 |
| Energy contribution | -20.34 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8488899 104 + 23771897 UGGUGGCUAAACCCUCUAUUCACCACUCCUUGGGUGGUGUGGUGGGAUCGGGAGAUUCAAAAUAAAC-ACCCCA--------CU--GGCGGCAUUUAUUGAUUUAUUUAUUGAUU ..((.((((..(((.(((..(((((((.....)))))))))).)))...(((...............-..))).--------.)--))).))....((..((......))..)). ( -29.23) >DroSec_CAF1 7327 112 + 1 UGGUGGCUAUACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGUAUUCAAAAUAAAC-ACCCCACCGACUGACU--GGCGGCAUUUAUUGAUUUAUUUAUUGAUU .((((...(((((((...(((((((((((......)))))))))))...)))))))..........)-)))((.(((......)--)).)).....((..((......))..)). ( -36.22) >DroSim_CAF1 7293 108 + 1 UGCUGGCUAAGCCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGUAUUCAAAAUAAAC-ACCCCACCGA----CU--GGCGGCAUUUAUUGAUUUAUUUAUUGAUU (((((.(...........(((((((((((......)))))))))))...(((((.............-))))).....----..--).)))))...((..((......))..)). ( -34.42) >DroEre_CAF1 8373 108 + 1 UGGUGGUUUGACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGUGAUCGGGGGAUUCAAAAUACAC-ACCCCACUGA----CU--GGCGGCAUUUAUUGAUUUAUUUAUUGAUU ..(((.((((((((((.((((((((((((......))))))))).))).)))))..)))))...)))-.(((((....----.)--)).)).....((..((......))..)). ( -37.10) >DroYak_CAF1 7349 97 + 1 GUGUG-----------UAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGCGGAUUCAAAAUACAC-ACCCCACCGA----CU--GGCGGCAUUUAUUGAUUUGUUUAUUGAUU (((((-----------(((((((((((((......)))))))))(((((....)))))..)))))))-)).((.((..----..--)).)).....((..((......))..)). ( -35.20) >DroAna_CAF1 7231 91 + 1 --GCAACCAAACC------UCACCAC-----------UGUGGGGA-AAAAGUCUUUUCAAAUAAAACUACCCCAUUAA----AUACGGAGCGUUUUAUUGAUUUUAAUAUUGAUU --.(((..(((((------((.....-----------.((((((.-...(((.(((......)))))).))))))...----.....))).))))..)))............... ( -15.04) >consensus UGGUGGCUAAACCCUCUAUUCACCACACCUUGGGUGGUGUGGUGGGAUCGGGGGAUUCAAAAUAAAC_ACCCCACCGA____CU__GGCGGCAUUUAUUGAUUUAUUUAUUGAUU ..((.(((..........(((((((((((......)))))))))))...((((................)))).............))).))....((..((......))..)). (-17.88 = -20.34 + 2.46)

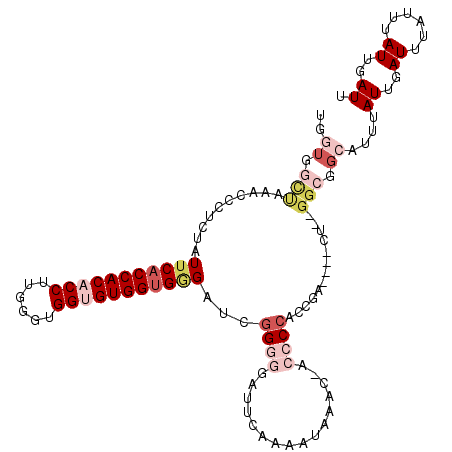

| Location | 8,488,899 – 8,489,003 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.85 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8488899 104 - 23771897 AAUCAAUAAAUAAAUCAAUAAAUGCCGCC--AG--------UGGGGU-GUUUAUUUUGAAUCUCCCGAUCCCACCACACCACCCAAGGAGUGGUGAAUAGAGGGUUUAGCCACCA ..........((((((((((((((((.((--..--------.)))))-)))))))......(((...((..((((((.((......)).)))))).)).)))))))))....... ( -25.80) >DroSec_CAF1 7327 112 - 1 AAUCAAUAAAUAAAUCAAUAAAUGCCGCC--AGUCAGUCGGUGGGGU-GUUUAUUUUGAAUACCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGUAUAGCCACCA .............(((.......((((.(--.....).))))(((((-((((.....))))))))))))..(((((((((......)))))))))....(..((.....))..). ( -38.00) >DroSim_CAF1 7293 108 - 1 AAUCAAUAAAUAAAUCAAUAAAUGCCGCC--AG----UCGGUGGGGU-GUUUAUUUUGAAUACCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGCUUAGCCAGCA .............(((.......((((..--..----.))))(((((-((((.....))))))))))))..(((((((((......)))))))))....(..(((...)))..). ( -38.20) >DroEre_CAF1 8373 108 - 1 AAUCAAUAAAUAAAUCAAUAAAUGCCGCC--AG----UCAGUGGGGU-GUGUAUUUUGAAUCCCCCGAUCACACCACACCACCCAAGGUGUGGUGAAUAGAGGGUCAAACCACCA ........................((((.--..----...))))(((-(.((...((((..(((.(.....(((((((((......)))))))))....).))))))))))))). ( -33.30) >DroYak_CAF1 7349 97 - 1 AAUCAAUAAACAAAUCAAUAAAUGCCGCC--AG----UCGGUGGGGU-GUGUAUUUUGAAUCCGCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUA-----------CACAC ........................(((((--..----..))))).((-(((((((....(((....)))..(((((((((......)))))))))))))-----------))))) ( -33.10) >DroAna_CAF1 7231 91 - 1 AAUCAAUAUUAAAAUCAAUAAAACGCUCCGUAU----UUAAUGGGGUAGUUUUAUUUGAAAAGACUUUU-UCCCCACA-----------GUGGUGA------GGUUUGGUUGC-- ..............(((((((((((((((((..----...))))))).)))))).))))...((((...-.((.(((.-----------...))).------))...))))..-- ( -21.40) >consensus AAUCAAUAAAUAAAUCAAUAAAUGCCGCC__AG____UCGGUGGGGU_GUUUAUUUUGAAUACCCCGAUCCCACCACACCACCCAAGGUGUGGUGAAUAGAGGGUUUAGCCACCA ..((((..(((((((......((.((((............)))).)).)))))))))))............(((((((((......))))))))).................... (-17.26 = -18.85 + 1.59)
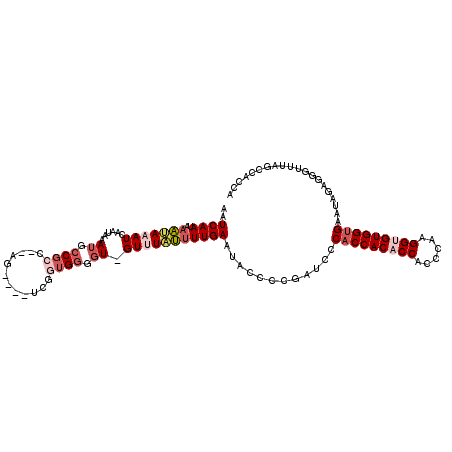

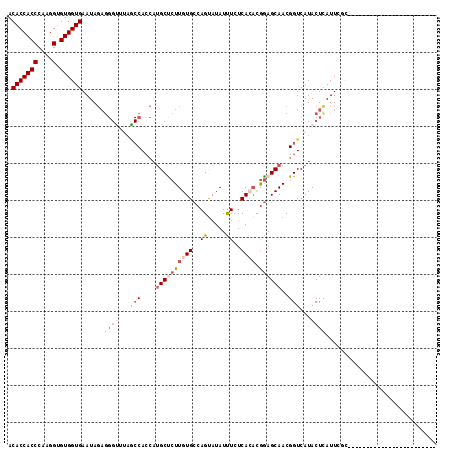
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:23 2006