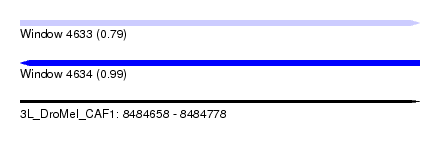
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,484,658 – 8,484,778 |
| Length | 120 |
| Max. P | 0.994736 |

| Location | 8,484,658 – 8,484,778 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
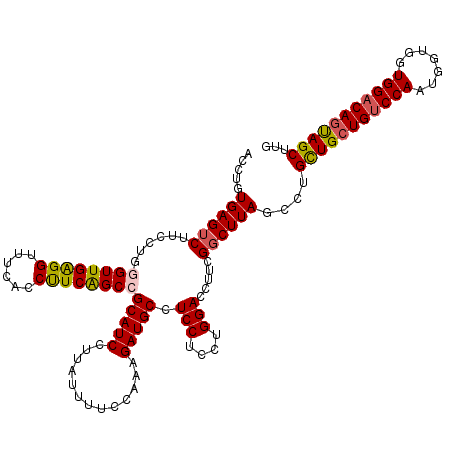
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -29.45 |
| Energy contribution | -30.97 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8484658 120 + 23771897 CAAGCUACUGUCCACAACCAUUGGACAGCAGCAGGCUAAGCCGAAGGUCCAGGAGGAGGCAUCUUUGGAAAAUAAGGAUGCGGCUGAAGGUGAAACCUCAACCCAGAAAGACUCACAGGU ...(((.(((((((.......))))))).))).(((...)))......((..(((...(((((((((.....)))))))))((.(((.((.....))))).))........)))...)). ( -38.50) >DroSec_CAF1 3111 120 + 1 CAAGCUACUGUCCACCACCAUUGGGCAGCAGCAGGCUAAGCCGAAGGUCCGGGAGGAGGCAUCUUUGGAAAAUAAGGAUGCGGCUGAAGGUGAAACCUCAACCCAGGAAGACUCACAGGU ...(((.(((((((.......))))))).))).(((...)))((.(.((((((..((((((((((((.....))))))))).((.....)).....)))..))).)))...)))...... ( -40.90) >DroSim_CAF1 3095 117 + 1 CAAGCUACUGUCCACCACCAUUGGACAGCAACAGGCUAAGCCGAAGGUCC---AGGAGGCAUCUUUGGAAAAUAAGGAUGCGGCUGAAGGUGAAACCUCAACCCAGGAAGACUCACAGGU ..((((.(((((((.......))))))).....))))..(((..((.(((---.....(((((((((.....)))))))))((.(((.((.....))))).))..)))...))....))) ( -35.10) >DroEre_CAF1 3346 120 + 1 UAAGCUAAUGUCCACCACCAUUGGACAGCAACAGGCUAAGCCCAAGGUCCAGGAGGAGGCAUCUGUGGAAAAGAACGAUGCGGCUGAGGAUGAAACCUCAACUCAGGAAGCCUCACCGGU ...(((...(((((.......))))))))...(((((.((((.....(((....))).(((((.((........)))))))))))((((......)))).........)))))....... ( -35.10) >DroYak_CAF1 3137 120 + 1 UAAGCUGCUGUCCACCACCAUUGGACAGCAGCAGGCUAAGCCAAAGGUCCAGGAGGAGGCAUCCUUGGAAAAGAAGGAUGCGGCCGAAGGUGAAACCCCAACUCAGAAAGACUCACAGGU ...(((((((((((.......))))))))))).((((..........(((....))).((((((((.......))))))))))))....((((...................)))).... ( -43.61) >consensus CAAGCUACUGUCCACCACCAUUGGACAGCAGCAGGCUAAGCCGAAGGUCCAGGAGGAGGCAUCUUUGGAAAAUAAGGAUGCGGCUGAAGGUGAAACCUCAACCCAGGAAGACUCACAGGU ...(((.(((((((.......))))))).)))..(((.((((.....(((....))).(((((((((.....)))))))))))))....((((...................)))).))) (-29.45 = -30.97 + 1.52)

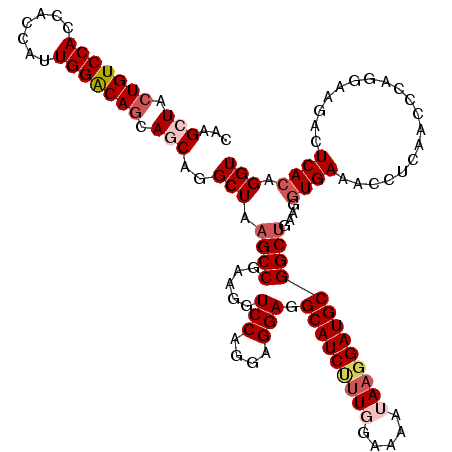
| Location | 8,484,658 – 8,484,778 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -36.54 |
| Energy contribution | -36.66 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8484658 120 - 23771897 ACCUGUGAGUCUUUCUGGGUUGAGGUUUCACCUUCAGCCGCAUCCUUAUUUUCCAAAGAUGCCUCCUCCUGGACCUUCGGCUUAGCCUGCUGCUGUCCAAUGGUUGUGGACAGUAGCUUG .....((((((......((((((((......))))))))(((((.............))))).(((....))).....))))))....(((((((((((.......)))))))))))... ( -41.62) >DroSec_CAF1 3111 120 - 1 ACCUGUGAGUCUUCCUGGGUUGAGGUUUCACCUUCAGCCGCAUCCUUAUUUUCCAAAGAUGCCUCCUCCCGGACCUUCGGCUUAGCCUGCUGCUGCCCAAUGGUGGUGGACAGUAGCUUG ..(((.(((........((((((((......))))))))(((((.............)))))...))).)))......(((...))).(((((((.(((.......))).)))))))... ( -36.72) >DroSim_CAF1 3095 117 - 1 ACCUGUGAGUCUUCCUGGGUUGAGGUUUCACCUUCAGCCGCAUCCUUAUUUUCCAAAGAUGCCUCCU---GGACCUUCGGCUUAGCCUGUUGCUGUCCAAUGGUGGUGGACAGUAGCUUG .((.(.(((........((((((((......))))))))(((((.............))))))))).---))......(((...))).(((((((((((.......)))))))))))... ( -38.92) >DroEre_CAF1 3346 120 - 1 ACCGGUGAGGCUUCCUGAGUUGAGGUUUCAUCCUCAGCCGCAUCGUUCUUUUCCACAGAUGCCUCCUCCUGGACCUUGGGCUUAGCCUGUUGCUGUCCAAUGGUGGUGGACAUUAGCUUA .((((.((((......(.(((((((......))))))))(((((.............)))))..)))))))).....((((...))))((((.((((((.......)))))).))))... ( -41.02) >DroYak_CAF1 3137 120 - 1 ACCUGUGAGUCUUUCUGAGUUGGGGUUUCACCUUCGGCCGCAUCCUUCUUUUCCAAGGAUGCCUCCUCCUGGACCUUUGGCUUAGCCUGCUGCUGUCCAAUGGUGGUGGACAGCAGCUUA ..............((((((..(((.((((.....((..((((((((.......))))))))..))...)))).)))..))))))...(((((((((((.......)))))))))))... ( -47.00) >consensus ACCUGUGAGUCUUCCUGGGUUGAGGUUUCACCUUCAGCCGCAUCCUUAUUUUCCAAAGAUGCCUCCUCCUGGACCUUCGGCUUAGCCUGCUGCUGUCCAAUGGUGGUGGACAGUAGCUUG .....((((((......((((((((......))))))))(((((.............))))).(((....))).....))))))....(((((((((((.......)))))))))))... (-36.54 = -36.66 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:16 2006