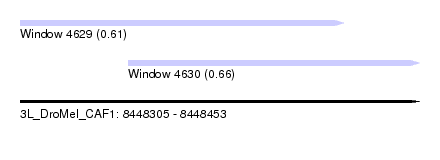
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,448,305 – 8,448,453 |
| Length | 148 |
| Max. P | 0.664239 |

| Location | 8,448,305 – 8,448,425 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -49.97 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8448305 120 + 23771897 AAACUCGCACCGGAGCACGCAUGCUGGCAUAGCCAAGUCGAGGAUUGGAUAAUCCAAAGCCAGGUUGAGUGACAUAGCCAUCCGACAGGGAAUGCUCACUGUCGGUGGUGUUGGCCAGAC ...((.(((((((..(((.((..(((((....((.......)).((((.....)))).)))))..)).)))......))..(((((((.((....)).))))))).))))).))...... ( -40.40) >DroPse_CAF1 3599 120 + 1 AUACCCUCACAGGGGCACGCAUACUGGCAUAGCCGAGGCGCGGAUUGGCAAAUCCAAGGCCCGGCUGUGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUGGUGUUGGCCAGGC ...(((.....)))....((......(((((((((.(((..(((((....)))))...))))))))))))(((...(((((((.(((((((....))))))))))))))....)))..)) ( -60.00) >DroGri_CAF1 6551 120 + 1 AGACGCGCACGGGUCCACGCAAGCUGGCAUAGCCGAGACGUGGAGCGGGCUGGCUGCUGCCCGCCUGCGUUGCAUAGCCAUCGGACAGCGAGUGCUCGCUAUCGGUGGUGUUGGCCAGAC .(((.(....).)))..(....)(((((.((((.(..(((..(.((((((........)))))))..)))..)...((((((((..(((((....))))).))))))))))))))))).. ( -55.40) >DroWil_CAF1 1386 120 + 1 AAACUCUCACCGGAGCACGCAUACUGGCAUAACCAAGACGAGGAUUAGACAUUCCGCUGCCCGGCUGAGUGGCAUAGCCAUCGGACAAAGAUUGCUCGCUGUCGGUGGUAUUGGCCAAGC .....((((((((.(((.((....(((.....)))......(((........)))))))))))).))))((((...(((((((.(((..((....))..))))))))))....))))... ( -42.50) >DroAna_CAF1 1266 120 + 1 AAACCCUCACCGGAGCACGCAUGCUGGCAUAACCCAGACGGGGAUUGGAGAGUCCACCUGCUGACUGGGUGGCGUAGCCAUCGGAGAGCGAGUGCUCGCUAUCGGUGGUGUUGGCCAGGC ....(((..(((..((((....((((((...((((((.(((.(..(((.....)))..).))).)))))).)).))))((((((..(((((....))))).)))))))))))))..))). ( -46.40) >DroPer_CAF1 3574 120 + 1 AUACCCUCACAGGGGCACGCAUACUGGCAUAGCCGAGGCGCGGAUUGGCAAAUCCAAGGCCCGGCUGUGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUAGUGUUGGCCAGGC ...(((.....))).........((((((((((((.(((..(((((....)))))...))))))))))..(((((.(((....((((((((....))))))))))).))))).))))).. ( -55.10) >consensus AAACCCUCACCGGAGCACGCAUACUGGCAUAGCCGAGACGAGGAUUGGAAAAUCCAAGGCCCGGCUGAGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUGGUGUUGGCCAGGC ...(((......)))........(((((...((..((.((.((..(((.....)))...)))).))..))..((..((((((.((((((((....))))))))))))))..))))))).. (-32.12 = -32.60 + 0.48)

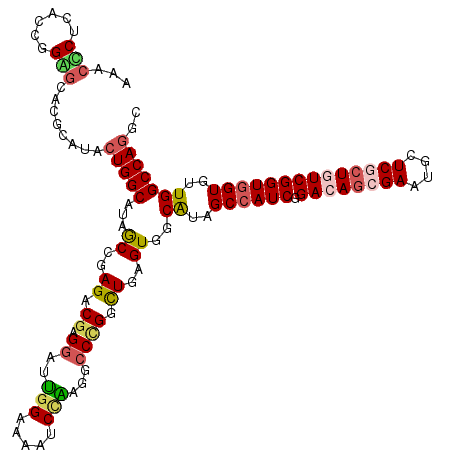
| Location | 8,448,345 – 8,448,453 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -28.41 |
| Energy contribution | -28.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8448345 108 + 23771897 AGGAUUGGAUAAUCCAAAGCCAGGUUGAGUGACAUAGCCAUCCGACAGGGAAUGCUCACUGUCGGUGGUGUUGGCCAGACUGGAGUCCAGUGCGC---CACUUCCCACCC---------C .(.(((((((..((((..((((((((....)))...((((.(((((((.((....)).))))))))))).))))).....))))))))))).)..---............---------. ( -37.50) >DroPse_CAF1 3639 120 + 1 CGGAUUGGCAAAUCCAAGGCCCGGCUGUGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUGGUGUUGGCCAGGCUGGAGUCCAAGCCGCCUCCAAUUCCUCCACCUCCUGGUCC .(((((....)))))..((((.(((((((...)))))))....((((((((....))))))))(((((.(((((..((((.((........)))))))))))....)))))....)))). ( -53.40) >DroWil_CAF1 1426 102 + 1 AGGAUUAGACAUUCCGCUGCCCGGCUGAGUGGCAUAGCCAUCGGACAAAGAUUGCUCGCUGUCGGUGGUAUUGGCCAAGCUGGAAUCUAGCCCGC---------CUCCAC---------C .(((........)))((((.((((((...((((...(((((((.(((..((....))..))))))))))....))))))))))....))))....---------......---------. ( -33.60) >DroYak_CAF1 3467 108 + 1 UGGAUUGGAUAAACCAAAGCCAGGUUGAGUGACAUAGCCAUCCGACAGGGAAUGCUCACUGUCGGUGGUGUUGGCCAGACUGGAGUCCAAUGCGC---CACUUCCUGCAC---------C .(.(((((((...(((..((((((((....)))...((((.(((((((.((....)).))))))))))).))))).....))).))))))).)((---........))..---------. ( -35.90) >DroAna_CAF1 1306 108 + 1 GGGAUUGGAGAGUCCACCUGCUGACUGGGUGGCGUAGCCAUCGGAGAGCGAGUGCUCGCUAUCGGUGGUGUUGGCCAGGCUGGAGUCCAAGCCUG---UAACUCCUCCAC---------C .(((..((((.(.((((((.......)))))))((.((((((((..(((((....))))).))))))))....))((((((((...)).))))))---...)))))))..---------. ( -46.20) >DroPer_CAF1 3614 120 + 1 CGGAUUGGCAAAUCCAAGGCCCGGCUGUGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUAGUGUUGGCCAGGCUGGAGUCCAAGCCACCUCCAAUUCCUCCACCUCCUGAUCC .((((..(.........((((.(((((((...)))))))..(.((((((((....)))))))).).......))))(((.(((((...................)))))))).)..)))) ( -45.81) >consensus AGGAUUGGAAAAUCCAAAGCCCGGCUGAGUGGCAUAGCCAUCGGACAGCGAAUGCUCGCUGUCGGUGGUGUUGGCCAGGCUGGAGUCCAAGCCGC___CAAUUCCUCCAC_________C .(((((((.....)).....((((((...((((...((((((.(((((.((....)).)))))))))))....)))))))))))))))................................ (-28.41 = -28.97 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:12 2006