
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,422,694 – 8,422,814 |
| Length | 120 |
| Max. P | 0.970446 |

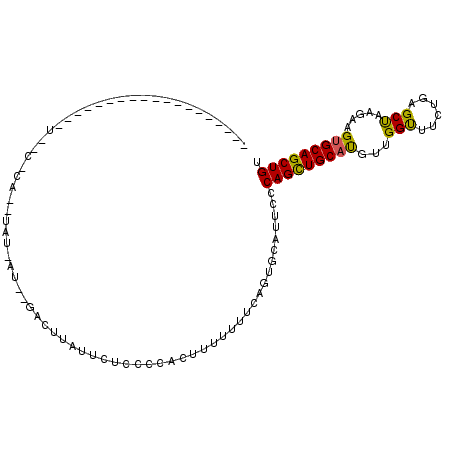
| Location | 8,422,694 – 8,422,784 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 63.25 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -16.32 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8422694 90 + 23771897 -------------------UAUCACAUAUAUAAUCAGACUUAUUUCCCCCACUUUUUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGCGCUAAGAAGUGCAGCUGU -------------------..............................((((((((...((((((...(((((.....)))))....))))))))))))))....... ( -19.60) >DroSec_CAF1 4638 69 + 1 ------------------------------------UACUU----UCCCCACUUUCUGUCAGUGCAUACCCAGCUGCACGUUGGUUUCUGAGCUAAGAAGUGCAGCUGU ------------------------------------.....----....((((.......))))......(((((((((.((((((....))))))...))))))))). ( -21.40) >DroSim_CAF1 11167 73 + 1 ------------------------------------UACUUAGUCACCCCACUUUCUUUCAGUGCAUUCCCAGCUGCAUUUCGGUUUCUGAGCUAAGAAGUGCAGCUGU ------------------------------------.............((((.......))))......((((((((((((((((....))))..)))))))))))). ( -21.60) >DroEre_CAF1 4712 109 + 1 CGAAAUAGAAUGUUCUGCAUGGCGCACUUAUUAUUCGAAUUGUUUGCCCAAACUUUUUUCAGUGCACUCCCAGCUGCAUGACGGCUUCCCAGCCAAGAAGUGCAGCUGU .....((((....))))...((.(((((........(((..((((....))))....)))))))).))..(((((((((...((((....)))).....))))))))). ( -28.00) >DroYak_CAF1 11276 96 + 1 -------------UCCGCAUACCACACCUAUUAUUUGACUUGUGCUCCCCAACUUUUUCCAGUGCAUUCACAGUUGCUUGAUGGUUUCCCAGCCCAGAAGUGCAGCUGU -------------.......................((..((..((..............))..)).))((((((((((..(((.........)))..)).)))))))) ( -16.84) >consensus ___________________U__C_CA__UAU_AU__GACUUAUUCUCCCCACUUUUUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGAGCUAAGAAGUGCAGCUGU ......................................................................(((((((((...(((......))).....))))))))). (-16.32 = -15.80 + -0.52)


| Location | 8,422,694 – 8,422,784 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 63.25 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -13.46 |
| Energy contribution | -14.06 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8422694 90 - 23771897 ACAGCUGCACUUCUUAGCGCAGAAACCAACAUGCAGCUGGGAAUGCACUGAAAAAAAGUGGGGGAAAUAAGUCUGAUUAUAUAUGUGAUA------------------- .(((.((((.(((((((((((..........))).)))))))))))))))......(((.(((........))).)))............------------------- ( -21.40) >DroSec_CAF1 4638 69 - 1 ACAGCUGCACUUCUUAGCUCAGAAACCAACGUGCAGCUGGGUAUGCACUGACAGAAAGUGGGGA----AAGUA------------------------------------ .(((((((((((((......))))......)))))))))....(.((((.......)))).)..----.....------------------------------------ ( -20.00) >DroSim_CAF1 11167 73 - 1 ACAGCUGCACUUCUUAGCUCAGAAACCGAAAUGCAGCUGGGAAUGCACUGAAAGAAAGUGGGGUGACUAAGUA------------------------------------ .(((.((((.(((((((((((..........)).))))))))))))))))..........((....)).....------------------------------------ ( -21.40) >DroEre_CAF1 4712 109 - 1 ACAGCUGCACUUCUUGGCUGGGAAGCCGUCAUGCAGCUGGGAGUGCACUGAAAAAAGUUUGGGCAAACAAUUCGAAUAAUAAGUGCGCCAUGCAGAACAUUCUAUUUCG .((((((((.....(((((....)))))...)))))))).(.(((((((.......(((((((.......)))))))....)))))))).................... ( -32.80) >DroYak_CAF1 11276 96 - 1 ACAGCUGCACUUCUGGGCUGGGAAACCAUCAAGCAACUGUGAAUGCACUGGAAAAAGUUGGGGAGCACAAGUCAAAUAAUAGGUGUGGUAUGCGGA------------- ....((((((((((.((((((....)).....(((........))).........)))).)))))((((..((........))))))...))))).------------- ( -23.70) >consensus ACAGCUGCACUUCUUAGCUCAGAAACCAACAUGCAGCUGGGAAUGCACUGAAAAAAAGUGGGGAAAACAAGUC__AU_AUA__UG_G__A___________________ .(((.((((.(((((((((...............))))))))))))))))........................................................... (-13.46 = -14.06 + 0.60)



| Location | 8,422,715 – 8,422,814 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.99 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -14.40 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8422715 99 + 23771897 UAUUUCCCCCACUUUUUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGCGCUAAGAAGUGCAGCUGU-------CCUGAUUUGCAGAAAAGGUCA---GCAAACAAC .........((((((((...((((((...(((((.....)))))....))))))))))))))..((((.-------(((............))).))---))....... ( -29.90) >DroSec_CAF1 4642 92 + 1 U----UCCCCACUUUCUGUCAGUGCAUACCCAGCUGCACGUUGGUUUCUGAGCUAAGAAGUGCAGCUGU-------CCUGAAUCGCAGAAAAAGUCA---GCAAAC--- .----.......(((((((.(...((....(((((((((.((((((....))))))...))))))))).-------..))..).)))))))......---......--- ( -26.70) >DroSim_CAF1 11171 96 + 1 UAGUCACCCCACUUUCUUUCAGUGCAUUCCCAGCUGCAUUUCGGUUUCUGAGCUAAGAAGUGCAGCUGU-------CCUGAAUCGCAGAAAAGGUCA---GCAAAC--- ..((.(((....(((((....(((.((((.((((((((((((((((....))))..)))))))))))).-------...)))))))))))).)))..---))....--- ( -27.40) >DroEre_CAF1 4752 96 + 1 UGUUUGCCCAAACUUUUUUCAGUGCACUCCCAGCUGCAUGACGGCUUCCCAGCCAAGAAGUGCAGCUGU-------GCUGAAUCGCAGAAAAGGUCA---GCAAAC--- .((((((....((((((((...........(((((((((...((((....)))).....)))))))))(-------((......)))))))))))..---))))))--- ( -32.00) >DroYak_CAF1 11303 96 + 1 UGUGCUCCCCAACUUUUUCCAGUGCAUUCACAGUUGCUUGAUGGUUUCCCAGCCCAGAAGUGCAGCUGU-------CCUGAAUCGCAGAAAAGGUCA---GCAAAG--- ..((((.....((((((((..(((.((((((((((((((..(((.........)))..)).))))))))-------...))))))).)))))))).)---)))...--- ( -27.20) >DroAna_CAF1 89178 95 + 1 ---CUACCCUAAUCUCUCUCAGUGUA-----AGCAGGACACUCG-UCCUUAUCUAA--AAUGCAGCUAUGCAGCCUUCUGAAUCCCUGAAAAGGUCAAAUGCCAAC--- ---...............((((....-----.(((((((....)-)))).......--..((((....))))))...))))...(((....)))............--- ( -18.20) >consensus U_UUUACCCCAAUUUCUUUCAGUGCAUUCCCAGCUGCAUGUUGGUUUCUGAGCUAAGAAGUGCAGCUGU_______CCUGAAUCGCAGAAAAGGUCA___GCAAAC___ ......................(((.....(((((((((...(((......))).....))))))))).........(((.....)))............)))...... (-14.40 = -15.23 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:57 2006