
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,412,871 – 8,412,980 |
| Length | 109 |
| Max. P | 0.548798 |

| Location | 8,412,871 – 8,412,980 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.64 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
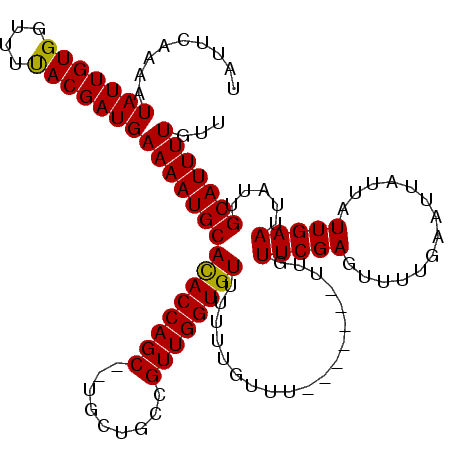
>3L_DroMel_CAF1 8412871 109 + 23771897 UAUUCAAAAUAUUGUGGUUUUACGAUGAAAAUGCACACCAGCUCUGCUGCCGUUGGUGUUU-UUAUUU-------UUGUUCGAGUUUUGAAUUAUUAUUGAAUUAUUGCAUUUUGUU .........(((((((....)))))))(((((((((((((((.........)))))))).(-(((...-------..((((((...))))))......)))).....)))))))... ( -21.50) >DroSim_CAF1 1372 108 + 1 UAUUCAAAAUAUUGUGGUUUUACGAUGAAAAUGCACACCAGC--UGCUGCCGUUGGUGUUUUUUGUUU-------UUGUUCGAGUUUUGAAUUAUUAUUGAAUUAUUGCAUUUUGUU .........(((((((....)))))))(((((((((((((((--.......)))))))).........-------..((((((..............))))))....)))))))... ( -21.84) >DroYak_CAF1 1388 115 + 1 UAUUCAAAAUAUUGUGGUUUCACGAUGAAAAUGCAUACCAGC--UGCUGCCGUUGGUGUUUUUUGUUUUUUUAUUUUUUUCGAGUUUUGAAUUAUUAUUGAAUUAUUGCAUUUUGUU .........(((((((....)))))))(((((((((((((((--.......))))))))...................(((((..............))))).....)))))))... ( -21.34) >consensus UAUUCAAAAUAUUGUGGUUUUACGAUGAAAAUGCACACCAGC__UGCUGCCGUUGGUGUUUUUUGUUU_______UUGUUCGAGUUUUGAAUUAUUAUUGAAUUAUUGCAUUUUGUU .........(((((((....)))))))(((((((((((((((.........))))))))...................(((((..............))))).....)))))))... (-20.08 = -19.64 + -0.44)

| Location | 8,412,871 – 8,412,980 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -13.80 |
| Consensus MFE | -11.73 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8412871 109 - 23771897 AACAAAAUGCAAUAAUUCAAUAAUAAUUCAAAACUCGAACAA-------AAAUAA-AAACACCAACGGCAGCAGAGCUGGUGUGCAUUUUCAUCGUAAAACCACAAUAUUUUGAAUA ..............((((((...............(((...(-------((((..-..((((((...((......))))))))..)))))..)))...............)))))). ( -14.21) >DroSim_CAF1 1372 108 - 1 AACAAAAUGCAAUAAUUCAAUAAUAAUUCAAAACUCGAACAA-------AAACAAAAAACACCAACGGCAGCA--GCUGGUGUGCAUUUUCAUCGUAAAACCACAAUAUUUUGAAUA ...(((((((................(((.......)))...-------.........((((((...((....--))))))))))))))).............(((....))).... ( -13.60) >DroYak_CAF1 1388 115 - 1 AACAAAAUGCAAUAAUUCAAUAAUAAUUCAAAACUCGAAAAAAAUAAAAAAACAAAAAACACCAACGGCAGCA--GCUGGUAUGCAUUUUCAUCGUGAAACCACAAUAUUUUGAAUA ...((((((((...............(((.......)))..........................((((....--))))...))))))))....(((....)))............. ( -13.60) >consensus AACAAAAUGCAAUAAUUCAAUAAUAAUUCAAAACUCGAACAA_______AAACAAAAAACACCAACGGCAGCA__GCUGGUGUGCAUUUUCAUCGUAAAACCACAAUAUUUUGAAUA ...(((((((................(((.......)))...................((((((...((......))))))))))))))).............(((....))).... (-11.73 = -12.07 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:52 2006