
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,398,207 – 8,398,457 |
| Length | 250 |
| Max. P | 0.963890 |

| Location | 8,398,207 – 8,398,297 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -16.76 |
| Energy contribution | -18.64 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8398207 90 - 23771897 CAUAUAAUUUCCGCCGUUUAUCGGCCGAUCCAGCAUGCCGGGCAUUAUUUAUAUAAUACUCGCACUGGUUUUGGUUGGCGAUCGGGAAAA .......(((((((((.....))))(((((((((..((((((((((((....)))))....)).)))))....)))))..))))))))). ( -29.80) >DroSec_CAF1 5500 90 - 1 CAUAUAAUUUCCGCCGUUUAUCGGCCGAUCCAUCAUGCCGGGCAUUAUUUAUAUAAUACUCGCACUGGUUUUGGUUGGCGAGCGGGUAAA .............((((((..(((((((.(((...(((.(((.(((((....))))).)))))).)))..)))))))..))))))..... ( -28.10) >DroSim_CAF1 5500 90 - 1 CAUAUAAUUUCCGCCGUUUAUCGGCCGAUCCAGCAUGCCGGGCAUUAUUUAUAUAAUACUCGCACUGGUUUUGGUUGGCGAGCGGGUAAA .............((((((..(((((((.((((..(((.(((.(((((....))))).))))))))))..)))))))..))))))..... ( -31.00) >DroEre_CAF1 3452 77 - 1 CAUAUAAUUUCCGCCGUUUAUCCAGCGAUCC------------AUUAUUUAUAUAAUACUCGCACUGGUU-UGCUUGGCGAGCGGGAAAA .......(((((((((((...(((((((...------------(((((....)))))..))))..)))..-.....)))).))))))).. ( -20.70) >DroYak_CAF1 3731 77 - 1 CAUAUAAUUUCCGCCGUUUAUCCAGCGAUCC------------AUUAUUUAUAUAAUACUCGCACUGGUU-UGCUAGUUGAGCGGGAAAG .......(((((((((........((((...------------(((((....)))))..))))(((((..-..))))))).))))))).. ( -19.90) >consensus CAUAUAAUUUCCGCCGUUUAUCGGCCGAUCCA_CAUGCCGGGCAUUAUUUAUAUAAUACUCGCACUGGUUUUGGUUGGCGAGCGGGAAAA .............((((((..((((((..(((......((((.(((((....))))).))))...)))...))))))..))))))..... (-16.76 = -18.64 + 1.88)


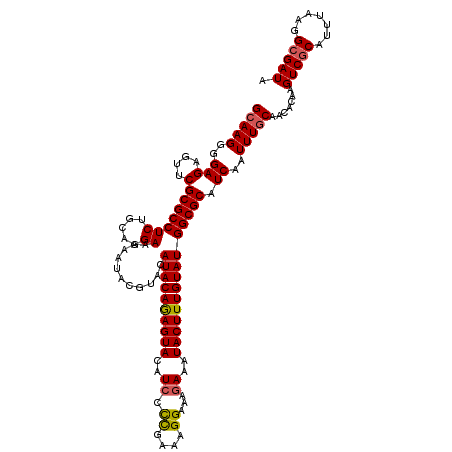
| Location | 8,398,297 – 8,398,417 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -27.69 |
| Energy contribution | -28.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8398297 120 + 23771897 GCAAGGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAAAGUAUAUCCUCAAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUCCAACACAAGUCGCAUUUAAGGCGAUA (((.((((...(....).))))))).(((((.((.(((.((((((((((((.((((....)))).....))))))))))))))).))...))))).......(((((.......))))). ( -36.30) >DroSec_CAF1 5590 120 + 1 GCAAGGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACAUCCCCGAAAGGAAACAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCCCAUUUAAGGCGAUA (((((..(((....)(((((((.....))...........((((((((((.....((....)).......))))))))))))))).))..))))).......(((((......)).))). ( -32.20) >DroSim_CAF1 5590 120 + 1 GCAAGGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACUUCCCUGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUA (((((..(((....)(((((((.....))...........((((((((((.(((.((....))...))).))))))))))))))).))..))))).......(((((.......))))). ( -34.30) >consensus GCAAGGGGAGAGUUCGCGCCUCUGCAGGAAAAAUACGUACAUACAGAGUACAUCCCCGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUA (((((..(((....)(((((((.....))...........((((((((((..(((......)))......))))))))))))))).))..))))).......(((((.......))))). (-27.69 = -28.13 + 0.45)


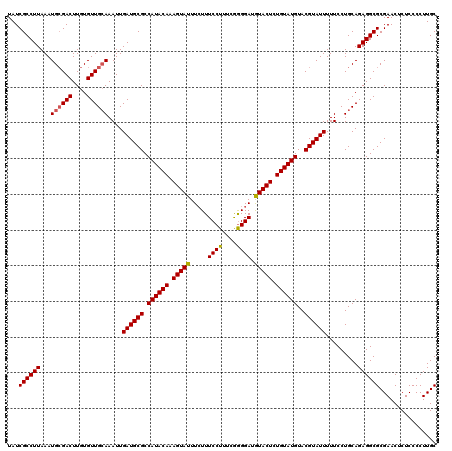
| Location | 8,398,297 – 8,398,417 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -30.54 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8398297 120 - 23771897 UAUCGCCUUAAAUGCGACUUGUGUUGGAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUUGAGGAUAUACUUUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCCUUGC ............((((.(((.(((.(((((..((((((.((((((((((((.(((((......))))).))))))))))))..))))))))))).))).))))))).............. ( -40.30) >DroSec_CAF1 5590 120 - 1 UAUCGCCUUAAAUGGGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUGUUUCCUUUCGGGGAUGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCCUUGC .............((((.((((((((((....((((((.((((((.(((((..((..((....))..))))))).))))))..)))))).....))))...))))))....))))..... ( -34.10) >DroSim_CAF1 5590 120 - 1 UAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCAGGGAAGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCCUUGC ...((((((...((((((....))))))....((((((.((((((.(((((((((((......))))))))))).))))))..)))))).........))))))................ ( -37.80) >consensus UAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCGGGGAUGUACUCUGUAUGUACGUAUUUUUCCUGCAGAGGCGCGAACUCUCCCCUUGC ...((((((...((((((....))))))....((((((.((((((.(((((.....((((....)))).))))).))))))..)))))).........))))))................ (-30.54 = -30.77 + 0.23)

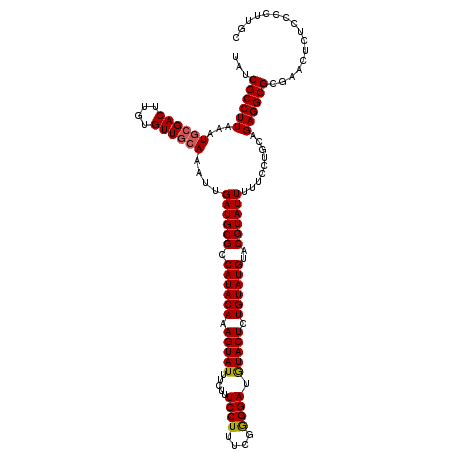
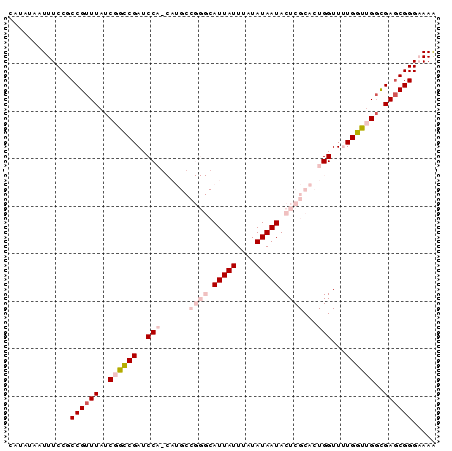
| Location | 8,398,337 – 8,398,457 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8398337 120 + 23771897 AUACAAAGUAUAUCCUCAAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUCCAACACAAGUCGCAUUUAAGGCGAUAGUUGAUCGAUCAUCUAAGACUUAAAAUUAAGUUGAUGUAU (((((((((((.((((....)))).....)))))))))))...((((((((((.......(((((((.......))..(((.(((....))).))).)))))......)))))))))).. ( -30.22) >DroSec_CAF1 5630 116 + 1 AUACAGAGUACAUCCCCGAAAGGAAACAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCCCAUUUAAGGCGAUAGUU----GAUCAUCAAAGACUUUAAAUUAAAUUGAUGUAU ((((((((((.....((....)).......))))))))))...(((((((((((......(((((((......)).(((....----.)))......))))).....))))))))))).. ( -28.50) >DroSim_CAF1 5630 116 + 1 AUACAGAGUACUUCCCUGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUC----GAUCAUCUAAGACUUUAAAUUAAAUUGAUGUAU ((((((((((.(((.((....))...))).))))))))))...(((((((((((........(((((.......)))))((((----..........))))......))))))))))).. ( -31.50) >consensus AUACAGAGUACAUCCCCGAAAGGAAAGAAAUACUUUGUAUGGCGCAUCAAUUUGCAACACAAGUCGCAUUUAAGGCGAUAGUU____GAUCAUCUAAGACUUUAAAUUAAAUUGAUGUAU ((((((((((..(((......)))......))))))))))...(((((((((((......(((((((.......))(((.(((....))).)))...))))).....))))))))))).. (-25.66 = -25.67 + 0.01)


| Location | 8,398,337 – 8,398,457 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8398337 120 - 23771897 AUACAUCAACUUAAUUUUAAGUCUUAGAUGAUCGAUCAACUAUCGCCUUAAAUGCGACUUGUGUUGGAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUUGAGGAUAUACUUUGUAU ...((((((.(((((..((((((.(((.(((....))).)))..((.......)))))))).)))))...))))))....(((((((((((.(((((......))))).))))))))))) ( -28.60) >DroSec_CAF1 5630 116 - 1 AUACAUCAAUUUAAUUUAAAGUCUUUGAUGAUC----AACUAUCGCCUUAAAUGGGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUGUUUCCUUUCGGGGAUGUACUCUGUAU ...((((((((((((...(((((...((((...----...)))).((......)))))))..))))..))))))))....(((((.(((((..((..((....))..))))))).))))) ( -28.00) >DroSim_CAF1 5630 116 - 1 AUACAUCAAUUUAAUUUAAAGUCUUAGAUGAUC----GACUAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCAGGGAAGUACUCUGUAU ...(((((((((.((((((.((..(((......----..)))..)).))))))(((((....))))))))))))))....(((((.(((((((((((......))))))))))).))))) ( -32.50) >consensus AUACAUCAAUUUAAUUUAAAGUCUUAGAUGAUC____AACUAUCGCCUUAAAUGCGACUUGUGUUGCAAAUUGAUGCGCCAUACAAAGUAUUUCUUUCCUUUCGGGGAUGUACUCUGUAU ...(((((((((......(((((...((((..........))))((.......))))))).......)))))))))....(((((.(((((.....((((....)))).))))).))))) (-20.99 = -21.22 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:45 2006