
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,393,607 – 8,393,765 |
| Length | 158 |
| Max. P | 0.988000 |

| Location | 8,393,607 – 8,393,725 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -27.71 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393607 118 + 23771897 A--UGUGUAUAUGCAUGCGAGCAUAUAUCACAGCAUCCAAAACUAAUUGAUCUUUCACCCAUCAAUCUCCGAUGAAGUGCGGUUAUCAAUUAGCCAAACGAUCGCGCUUACAAGUCAAAU .--(((((((((((......))))))).))))..............(((((........((((.......))))((((((((((...............))))))))))....))))).. ( -27.96) >DroSec_CAF1 824 120 + 1 AUAUGUAUAUAUGCAUGCGAGCAUAUAUCACAGCAUCCAAAACUAAUUGAUCUUUCACCCAUCAAUCUCCGAUGAAGUGCGGUUAUCAAUUAGCCAAAUGAUCGCGCUUACAAGUCAAAC ...(((((((((((......)))))))).)))..............(((((........((((.......))))((((((((((((...........))))))))))))....))))).. ( -28.40) >DroSim_CAF1 808 120 + 1 AUAUGUAUGUAUGCAUGCGAGCAUAUAUCACAGCAUCCAAAACUAAUUGAUCUUUCACCCAUCAAUCUCCGAUGAAGUGCGGUUAUCAAUUAGCCAAAUGAUCGCGCUUACAAGUCAAAC ...(((((((((((......)))))))).)))..............(((((........((((.......))))((((((((((((...........))))))))))))....))))).. ( -28.40) >consensus AUAUGUAUAUAUGCAUGCGAGCAUAUAUCACAGCAUCCAAAACUAAUUGAUCUUUCACCCAUCAAUCUCCGAUGAAGUGCGGUUAUCAAUUAGCCAAAUGAUCGCGCUUACAAGUCAAAC ...(((((((((((......)))))))).)))..............(((((........((((.......))))((((((((((((...........))))))))))))....))))).. (-27.71 = -27.60 + -0.11)


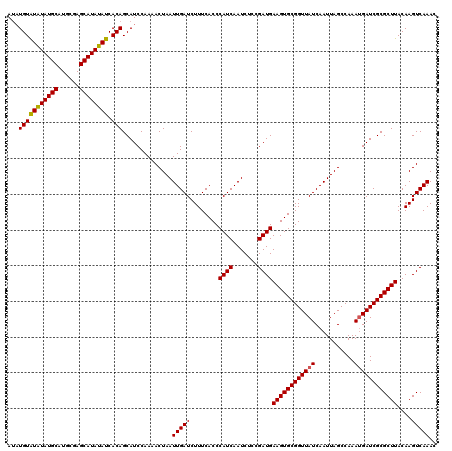
| Location | 8,393,607 – 8,393,725 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -34.01 |
| Energy contribution | -33.90 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393607 118 - 23771897 AUUUGACUUGUAAGCGCGAUCGUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUUUUGGAUGCUGUGAUAUAUGCUCGCAUGCAUAUACACA--U ...............(((.(((...(((((((((((.....((((.(((((.....)))))))))....))))))))))).))).)))((((.(((((((......)))))))))))--. ( -35.70) >DroSec_CAF1 824 120 - 1 GUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUUUUGGAUGCUGUGAUAUAUGCUCGCAUGCAUAUAUACAUAU (((((.....)))))(((.(((...(((((((((((.....((((.(((((.....)))))))))....))))))))))).))).)))((((((((((((......)))))))).)))). ( -37.20) >DroSim_CAF1 808 120 - 1 GUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUUUUGGAUGCUGUGAUAUAUGCUCGCAUGCAUACAUACAUAU (((((.....)))))(((.(((...(((((((((((.....((((.(((((.....)))))))))....))))))))))).))).)))((((((.(((((......))))).)).)))). ( -33.10) >consensus GUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUUUUGGAUGCUGUGAUAUAUGCUCGCAUGCAUAUAUACAUAU ...............(((.(((...(((((((((((.....((((.(((((.....)))))))))....))))))))))).))).)))((((.(((((((......)))))))))))... (-34.01 = -33.90 + -0.11)



| Location | 8,393,645 – 8,393,765 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -32.27 |
| Energy contribution | -32.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393645 120 - 23771897 GGGUUACAUCGUUUAAAGUUGACCCGAAGCGAAAUGAUUUAUUUGACUUGUAAGCGCGAUCGUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUU ((((((...(.......).)))))).((((((..((.(((((.......)))))))...))))))(((((((((((.....((((.(((((.....)))))))))....))))))))))) ( -30.00) >DroSec_CAF1 864 120 - 1 GGGUUACAUCGUUUAAAGUUGACCCGGAGUGAAAUGAUUUGUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUU ((((((...(.......).))))))......((((((((((((((.....)))))).))))))))(((((((((((.....((((.(((((.....)))))))))....))))))))))) ( -33.30) >DroSim_CAF1 848 120 - 1 GGGUUACAUCGUUUAAAGUUGACCCGGAGCGAAAUGAUUUGUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUU ((((((...(.......).))))))..(((.((((((((((((((.....)))))).)))))))).)))(((((((.....((((.(((((.....)))))))))....))))))).... ( -34.10) >consensus GGGUUACAUCGUUUAAAGUUGACCCGGAGCGAAAUGAUUUGUUUGACUUGUAAGCGCGAUCAUUUGGCUAAUUGAUAACCGCACUUCAUCGGAGAUUGAUGGGUGAAAGAUCAAUUAGUU ((((((...(.......).))))))...((.((((((((((((((.....)))))).)))))))).))((((((((.....((((.(((((.....)))))))))....))))))))... (-32.27 = -32.17 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:33 2006