
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,393,168 – 8,393,407 |
| Length | 239 |
| Max. P | 0.914898 |

| Location | 8,393,168 – 8,393,287 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -28.01 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393168 119 - 23771897 GGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUGCACUGGGAGAAAUCAGCAGAACAUAUGAACAAAAGGAAAUCAGUAAAUCAGAGGUGUAUUU (((.(((....(((((....)))))(((.....)))))).)))....((((((((((...(((.((....))..)))......(((...........))).........)))))))))) ( -37.70) >DroSec_CAF1 400 116 - 1 GGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCAUCUGCACUGGGAGAAAUCAGCAGAACAUAUGAAGAAAAGUUA---GGCGAAUCAGAGGACUUUUU (((.(((....(((((....)))))(((.....)))))).)))......(((..(((((.....((....)).))))).))).....((((((((.---..(......)..)))))))) ( -35.30) >DroSim_CAF1 389 116 - 1 GGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUACACUGGGAGAAAUCAGCGGAACAUAUGAACAAAAGUUA---AGCAAAUCAGAGGACUUUUU (((.(((....(((((....)))))(((.....)))))).)))...........(((...((((((....)).((..(((............))).---.))...)))))))....... ( -31.90) >consensus GGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUGCACUGGGAGAAAUCAGCAGAACAUAUGAACAAAAGUUA___AGCAAAUCAGAGGACUUUUU (((.(((....(((((....)))))(((.....)))))).))).......(((.((......))((....)).)))........................................... (-28.01 = -27.57 + -0.44)


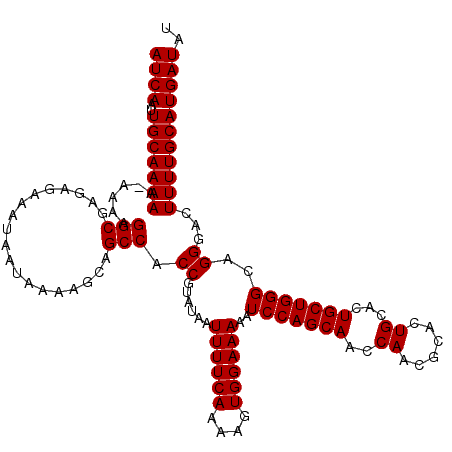
| Location | 8,393,208 – 8,393,327 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -34.49 |
| Energy contribution | -34.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393208 119 + 23771897 UUCUGCUGAUUUCUCCCAGUGCAGGUGCAUUUUCCUGGCCUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAAAAAAAGGCGAGAGAAAUAAUAAAAGC .........(((((((((((((....)))))....((((.((((((.....)))(((((....)))))....))).))))..................)).))))))............ ( -35.00) >DroSec_CAF1 437 118 + 1 UUCUGCUGAUUUCUCCCAGUGCAGAUGCAUUUUCCUGGCCUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAA-AAAAGGCGAGAGAAAUAAUAAAAGC .........(((((((((((((....)))))....((((.((((((.....)))(((((....)))))....))).)))).............-....)).))))))............ ( -35.00) >DroSim_CAF1 426 117 + 1 UUCCGCUGAUUUCUCCCAGUGUAGGUGCAUUUUCCUGGCCUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAA--AAAAGGCGAGAGAAAUAAUAAAAGC .........((((((((..(.....((((......((((.((((((.....)))(((((....)))))....))).)))).....))))...--.)..)).))))))............ ( -34.80) >consensus UUCUGCUGAUUUCUCCCAGUGCAGGUGCAUUUUCCUGGCCUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAA_AAAAGGCGAGAGAAAUAAUAAAAGC .........(((((((((((((....)))))....((((.((((((.....)))(((((....)))))....))).))))..................)).))))))............ (-34.49 = -34.27 + -0.22)



| Location | 8,393,208 – 8,393,327 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -37.40 |
| Energy contribution | -37.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393208 119 - 23771897 GCUUUUAUUAUUUCUCUCGCCUUUUUUUUUUGCAAUUGAUGGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUGCACUGGGAGAAAUCAGCAGAA (((......((((((((((((...................))).......(((((((.((((.(((((.....)))))))))((......))..))))).))))))))))).))).... ( -38.71) >DroSec_CAF1 437 118 - 1 GCUUUUAUUAUUUCUCUCGCCUUUU-UUUUUGCAAUUGAUGGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCAUCUGCACUGGGAGAAAUCAGCAGAA (((......((((((((((......-....((((.((..((((.(((....(((((....)))))(((.....)))))).))))...)).)))).......)))))))))).))).... ( -38.43) >DroSim_CAF1 426 117 - 1 GCUUUUAUUAUUUCUCUCGCCUUUU--UUUUGCAAUUGAUGGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUACACUGGGAGAAAUCAGCGGAA (((......((((((((((......--...((((.((..((((.(((....(((((....)))))(((.....)))))).))))...)).)))).......)))))))))).))).... ( -38.29) >consensus GCUUUUAUUAUUUCUCUCGCCUUUU_UUUUUGCAAUUGAUGGCAUCCGAUAGGCAGGAGCCUGCCAGCAAUUCGCUGGAGGCCAGGAAAAUGCACCUGCACUGGGAGAAAUCAGCAGAA (((......((((((((((...........((((.((..((((.(((....(((((....)))))(((.....)))))).))))...)).)))).......)))))))))).))).... (-37.40 = -37.40 + 0.00)



| Location | 8,393,247 – 8,393,367 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -25.91 |
| Energy contribution | -25.91 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393247 120 + 23771897 CUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAAAAAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAAC ..(((((.....)))))..((((((........))).))).....((((..........(((...................)))...................((((....)))))))). ( -25.91) >DroSec_CAF1 476 119 + 1 CUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAA-AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAAC ..(((((.....)))))..((((((........))).))).....((((.....-....(((...................)))...................((((....)))))))). ( -25.91) >DroSim_CAF1 465 118 + 1 CUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAA--AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAAC ..(((((.....)))))..((((((........))).))).....((((....--....(((...................)))...................((((....)))))))). ( -25.91) >consensus CUCCAGCGAAUUGCUGGCAGGCUCCUGCCUAUCGGAUGCCAUCAAUUGCAAAAA_AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAAC ..(((((.....)))))..((((((........))).))).....((((..........(((...................)))...................((((....)))))))). (-25.91 = -25.91 + 0.00)

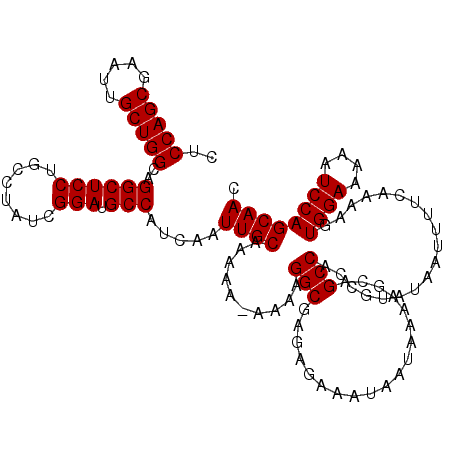

| Location | 8,393,287 – 8,393,407 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -24.84 |
| Energy contribution | -24.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8393287 120 + 23771897 AUCAAUUGCAAAAAAAAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAACCAACGCACUGCACUGCUGGGCAGGGACUUUUGCAUGAUAU ((((..(((((((......(((...................))).((......((((((....))))))..(((((((..((......))...)))))))..))...))))))))))).. ( -25.11) >DroSec_CAF1 516 119 + 1 AUCAAUUGCAAAAA-AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAACCAACGCACUGCACUGCUGGGCAGGGACUUUUGCAUGAUAU ((((..(((((((.-....(((...................))).((......((((((....))))))..(((((((..((......))...)))))))..))...))))))))))).. ( -25.11) >DroSim_CAF1 505 118 + 1 AUCAAUUGCAAAA--AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAACCAAUGCACUGCACUGCUGGGCAGGGACUUUUGCAUGAUAU ((((..(((((((--....(((...................))).((......((((((....))))))..(((((((.....(((...))).)))))))..))...))))))))))).. ( -25.31) >consensus AUCAAUUGCAAAAA_AAAAGGCGAGAGAAAUAAUAAAAGCAGCCACCGUAUAAUUUUCAAAAGUGGAAAAAUCCAGCAACCAACGCACUGCACUGCUGGGCAGGGACUUUUGCAUGAUAU ((((..(((((((......(((...................))).((......((((((....))))))..(((((((..((......))...)))))))..))...))))))))))).. (-24.84 = -24.84 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:31 2006