
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,392,808 – 8,393,078 |
| Length | 270 |
| Max. P | 0.989746 |

| Location | 8,392,808 – 8,392,918 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -44.63 |
| Energy contribution | -44.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392808 110 + 23771897 GAGCAGACGCAUUCUCAGCUUAAUGGACUCGGAUGCUUCUCUGGCUGGAUGCGAGUCCGAAUCUGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAA ..((....)).(((((.(((((((((.((((((((((((((((..(((((.((....)).)))).)..))))))....))))))).)))...)))))))))...))))). ( -46.90) >DroSec_CAF1 39 110 + 1 GAGCAGACGCAUUCUCAGCUUAAUGGACUCGGAUGCUUCUCUGGCUGGAUGCGAAUCCGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCACGGAGAAA ..((....)).(((((.(((((((((.((((((((((...(.....)(((((...(((((.......))))).)))))))))))).)))...)))))))))...))))). ( -46.50) >DroSim_CAF1 30 110 + 1 GAGCAGACGCAUUCUCAGCUUAAUGGACUCGGAUGCUUCUCUGGCUGGAUGCGAAUACGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAA ..((....)).(((((.(((((((((.((((((((((((((((.((((...((....))..))))...))))))....))))))).)))...)))))))))...))))). ( -45.50) >consensus GAGCAGACGCAUUCUCAGCUUAAUGGACUCGGAUGCUUCUCUGGCUGGAUGCGAAUCCGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAA ..((....)).(((((.(((((((((.((((((((((...(.....)(((((...(((((.......))))).)))))))))))).)))...)))))))))...))))). (-44.63 = -44.97 + 0.33)

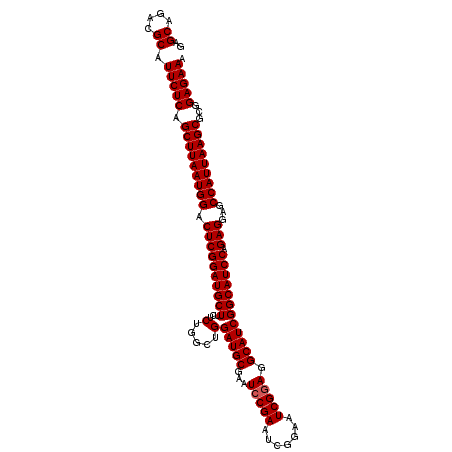

| Location | 8,392,808 – 8,392,918 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -39.50 |
| Energy contribution | -39.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392808 110 - 23771897 UUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUCCGAUUCAGAUUCGGACUCGCAUCCAGCCAGAGAAGCAUCCGAGUCCAUUAAGCUGAGAAUGCGUCUGCUC .(((((...((((((((((((((((((.((..(((((.(((((.......)))))...))))))).))))))........))).))))))))).))))).((....)).. ( -41.50) >DroSec_CAF1 39 110 - 1 UUUCUCCGUGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUCCGAUUCCGAUUCGGAUUCGCAUCCAGCCAGAGAAGCAUCCGAGUCCAUUAAGCUGAGAAUGCGUCUGCUC .(((((...((((((((((((((((((.((..(((((.(((((.......)))))...))))))).))))))........))).))))))))).))))).((....)).. ( -42.20) >DroSim_CAF1 30 110 - 1 UUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUCCGAUUCCGAUUCGUAUUCGCAUCCAGCCAGAGAAGCAUCCGAGUCCAUUAAGCUGAGAAUGCGUCUGCUC .(((((...((((((((((((..((((((((.(((((...((....))....))))).))))))))...((......)).))).))))))))).))))).((....)).. ( -37.70) >consensus UUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUCCGAUUCCGAUUCGGAUUCGCAUCCAGCCAGAGAAGCAUCCGAGUCCAUUAAGCUGAGAAUGCGUCUGCUC .(((((...((((((((((((((((((.((..(((((.(((((.......)))))...))))))).))))))........))).))))))))).))))).((....)).. (-39.50 = -39.83 + 0.33)

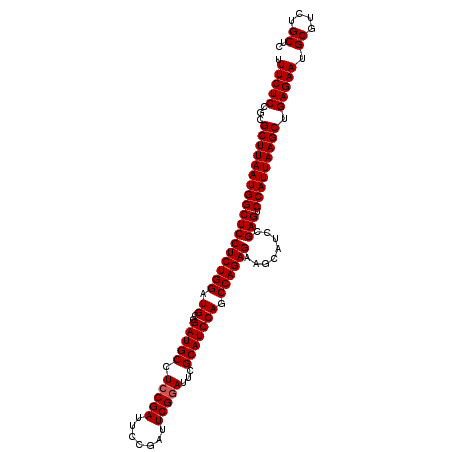

| Location | 8,392,838 – 8,392,958 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -38.10 |
| Energy contribution | -38.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392838 120 + 23771897 GGAUGCUUCUCUGGCUGGAUGCGAGUCCGAAUCUGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCC ((((((((((((((((.(((((...(((((.......))))).)))))(((.(((...))))))....))).))))))......))).(((((((...)))))))...........)))) ( -39.60) >DroSec_CAF1 69 120 + 1 GGAUGCUUCUCUGGCUGGAUGCGAAUCCGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCACGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCC ((((((((((((((((.(((((...(((((.......))))).)))))(((.(((...))))))....))).))))))......))).(((((((...)))))))...........)))) ( -40.70) >DroSim_CAF1 60 120 + 1 GGAUGCUUCUCUGGCUGGAUGCGAAUACGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCC ((((((((((((((((.(((((.....(((.......)))...)))))(((.(((...))))))....))).))))))......))).(((((((...)))))))...........)))) ( -35.10) >consensus GGAUGCUUCUCUGGCUGGAUGCGAAUCCGAAUCGGAAUCGGAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCC ((((((((((((((((.(((((...(((((.......))))).)))))(((.(((...))))))....))).))))))......))).(((((((...)))))))...........)))) (-38.10 = -38.43 + 0.33)


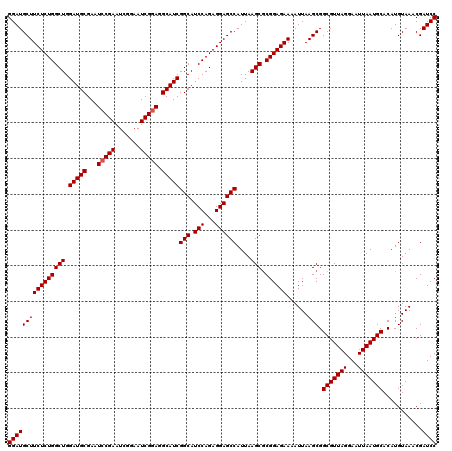
| Location | 8,392,878 – 8,392,998 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -36.42 |
| Energy contribution | -36.75 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392878 120 + 23771897 GAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAU ..(((.(((((.(((...)))))).......(((((............(((((((...)))))))............)))))..)).(((((...(((....)))..)))))..)))... ( -38.55) >DroSec_CAF1 109 120 + 1 GAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCACGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAGAGCCGAGACCACAGCCAAU ........(((.(((((....((......)).((((............(((((((...)))))))............))))))))).(((((...(((....)))..)))))..)))... ( -35.65) >DroSim_CAF1 100 120 + 1 GAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAU ..(((.(((((.(((...)))))).......(((((............(((((((...)))))))............)))))..)).(((((...(((....)))..)))))..)))... ( -38.55) >consensus GAGGCAUCGGCAUCCAGAGGAGCCAUUAAGCGCGGAGAAAAUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAU ..(((.(((((.(((...)))))).......(((((............(((((((...)))))))............)))))..)).(((((...(((....)))..)))))..)))... (-36.42 = -36.75 + 0.33)

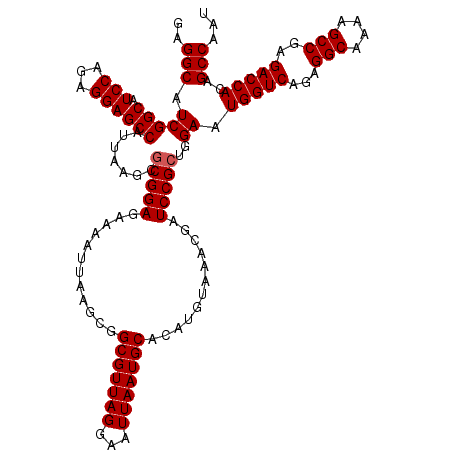

| Location | 8,392,878 – 8,392,998 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -36.31 |
| Consensus MFE | -36.16 |
| Energy contribution | -35.94 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392878 120 - 23771897 AUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAUUUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUC ((((((.((((((..(((....)))...))))))(((((.((((((((.....((((..............................))))..))))).))).))))).))))))..... ( -36.81) >DroSec_CAF1 109 120 - 1 AUUGGCUGUGGUCUCGGCUCUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAUUUUCUCCGUGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUC ((((((.((((((..(((....)))...))))))(((((.((((((((..((((...(((((...(....)....)))))......))))...))))).))).))))).))))))..... ( -35.30) >DroSim_CAF1 100 120 - 1 AUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAUUUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUC ((((((.((((((..(((....)))...))))))(((((.((((((((.....((((..............................))))..))))).))).))))).))))))..... ( -36.81) >consensus AUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAUUUUCUCCGCGCUUAAUGGCUCCUCUGGAUGCCGAUGCCUC ((((((.((((((..(((....)))...))))))(((((.((((((((.....((((..............................))))..))))).))).))))).))))))..... (-36.16 = -35.94 + -0.22)

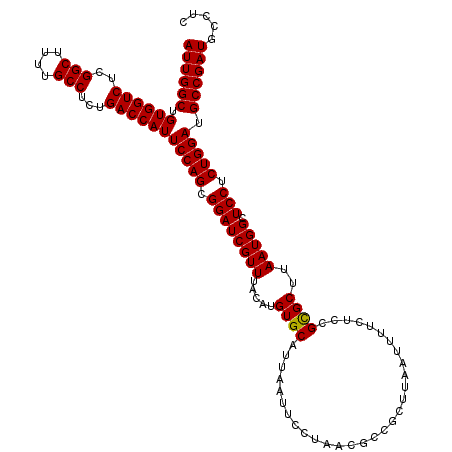

| Location | 8,392,918 – 8,393,038 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -34.20 |
| Energy contribution | -35.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392918 120 + 23771897 AUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAUCUCAUUCAGCAUUGCAGCAAUGCGACCUCAACCGCCCCAC .....(.(((((((((....(((((..(((.....(((..((((...(((((...(((....)))..)))))))))..))).)))...)))))(((....)))..))).)).)))).).. ( -35.70) >DroSec_CAF1 149 120 + 1 AUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAGAGCCGAGACCACAGCCAAUCUCAUUCAGCAUUGCAGCAAUGCGACCUCAACCGCCCCAC .....(.(((((((((....(((((..(((.....(((..((((...(((((...(((....)))..)))))))))..))).)))...)))))(((....)))..))).)).)))).).. ( -35.70) >DroSim_CAF1 140 120 + 1 AUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAUCUCAUCCAGCAUUGCAGCAAUGCGACCUCAACCGCCCCAC .....((((.(((.(((.....(((....)))...(((..((((...(((((...(((....)))..)))))))))..)))...)))))).(((((....)))))......))))..... ( -35.80) >consensus AUUAAGCGGCGUUAGGAAUUAAUGCACAUGUAAACGAUCCGCUGGAAUGGUCAGAGGCAAAAGCCGAGACCACAGCCAAUCUCAUUCAGCAUUGCAGCAAUGCGACCUCAACCGCCCCAC .....(.(((((((((....(((((..(((.....(((..((((...(((((...(((....)))..)))))))))..))).)))...)))))(((....)))..))).)).)))).).. (-34.20 = -35.70 + 1.50)


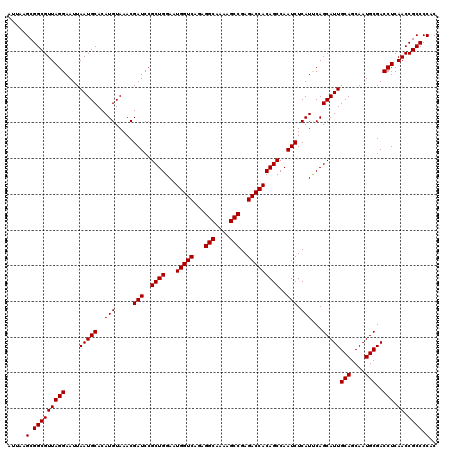
| Location | 8,392,918 – 8,393,038 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -44.49 |
| Energy contribution | -44.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.18 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392918 120 - 23771897 GUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAU ...(((((((..(((....((((.(((((((.(((((..((((.(((((((((..(((....)))...)))))...)))).)))))))))))).)))).)))).)))...)))))))... ( -44.40) >DroSec_CAF1 149 120 - 1 GUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUCUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAU ...(((((((..(((....((((.(((((((.(((((..((((.(((((((((..(((....)))...)))))...)))).)))))))))))).)))).)))).)))...)))))))... ( -44.40) >DroSim_CAF1 140 120 - 1 GUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGGAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAU ...(((((((..(((....((((.(((((((.(((((..((((.(((((((((..(((....)))...)))))...)))).)))))))))))).)))).)))).)))...)))))))... ( -44.00) >consensus GUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGCGGAUCGUUUACAUGUGCAUUAAUUCCUAACGCCGCUUAAU ...(((((((..(((....((((.(((((((.(((((..((((.(((((((((..(((....)))...)))))...)))).)))))))))))).)))).)))).)))...)))))))... (-44.49 = -44.27 + -0.22)



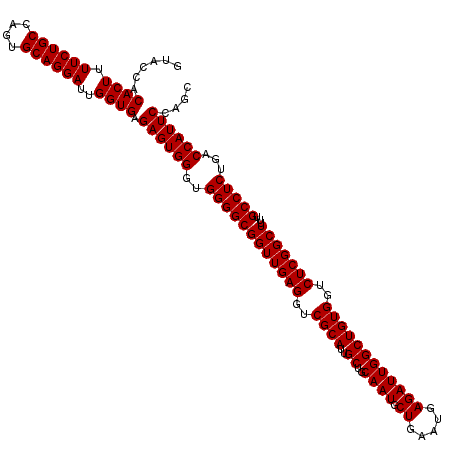
| Location | 8,392,958 – 8,393,078 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -46.10 |
| Energy contribution | -46.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8392958 120 - 23771897 GUACCACACUUUUCUGCCAGUGCAGGAUUGGUGAGAGUGGGUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGC ......((((.((((((....))))))..)))).((((((..((((((((((((..((((..((..((((.((.....))))))))))))..)))))))...)))))...)))))).... ( -46.10) >DroSec_CAF1 189 120 - 1 GUACCACACUUUUCUGCCAGUGCAGGAUUGGUGAGAGUGGGUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUCUUGCCUCUGACCAUUCCAGC ......((((.((((((....))))))..)))).((((((..((((((((((((..((((..((..((((.((.....))))))))))))..)))))))...)))))...)))))).... ( -46.10) >DroSim_CAF1 180 120 - 1 GUACCACACUUUUCUGCCAGUGCAGGAUUGGUGAGAGUGGGUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGGAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGC ......((((.((((((....))))))..)))).((((((..((((((((((((..((((..((..((((.((.....))))))))))))..)))))))...)))))...)))))).... ( -46.10) >consensus GUACCACACUUUUCUGCCAGUGCAGGAUUGGUGAGAGUGGGUGGGGCGGUUGAGGUCGCAUUGCUGCAAUGCUGAAUGAGAUUGGCUGUGGUCUCGGCUUUUGCCUCUGACCAUUCCAGC ......((((.((((((....))))))..)))).((((((..((((((((((((..((((..((..((((.((.....))))))))))))..)))))))...)))))...)))))).... (-46.10 = -46.10 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:26 2006