| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 831,563 – 831,659 |
| Length | 96 |
| Max. P | 0.909022 |

| Location | 831,563 – 831,659 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -17.06 |
| Energy contribution | -19.45 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 831563 96 + 23771897 GAAUUGGGUGAGUUGAGGAUGAGCCAGGCCACCGACACCGAGCACUGGAGCACCAUCCAGGCCUUGUUCGGUGAAGGAGAGC--CAAGAAGUCAACUC-------- .........(((((((((.....)).((((.((..((((((((((((((......)))))....)))))))))..)).).))--)......)))))))-------- ( -39.20) >DroPse_CAF1 6770 105 + 1 GAGCUGUCCGAGCUGAGGGUGCGACAAUCUACAGAUUCGGAGCACUGGCAGACCAUACACGCUCUGUUCGGUGAACAG-GGCACUGGGGAGACGACUCCCCGUGCC ...((((((((((.((((((((..(((((....)))).)..)))))((....)).......))).))))))...))))-(((((.((((((....))))))))))) ( -47.30) >DroSec_CAF1 6051 96 + 1 GAAUUGGGUGAGUUGAGGAUGAGCCAGGCCACCGACACCGAGCACUGGAGCACCAUCCAGGCCUUGUUCGGCGAUGGAGAGC--AAAGAAGUCAACUC-------- .........(((((((((.....))..(((.(((.(.((((((((((((......)))))....))))))).).))).).))--.......)))))))-------- ( -31.90) >DroSim_CAF1 5479 95 + 1 GAAUUGGGUGAGUUGAGGAUGAGCCAGGCCACCGACACCGAGCACUGGAGCACCAUCCAGGCCUUGUUCGGUGAUGGA-AGC--AAAGGAGUCAACUC-------- .........(((((((((.....))..((..(((.((((((((((((((......)))))....))))))))).))).-.))--.......)))))))-------- ( -35.40) >DroEre_CAF1 6898 96 + 1 GAGUUGGGAGAGUUGCGGAUAAGCCAGGCCACCGACACCGAGCACUGGAGCACCAUUCAAGCCUUGUUCGGUGACGGAGGGC--CAAGAAGGCAACCC-------- ...........(((((((.....)).((((.(((.(((((((((..((.((.........))))))))))))).)))..)))--)......)))))..-------- ( -37.30) >DroAna_CAF1 6486 96 + 1 GAGUUAAAUGAACUGCGACUUCGUCAGGCCUCAGAAACCGAGCACUGGCAGACCAUUCAGGCCUUGUUCGGUGAAGGAUCUA--AUGAAAUUCAACAU-------- (((((.......(((((....)).))).(((.....((((((((..(((.((....))..))).))))))))..))).....--....))))).....-------- ( -25.60) >consensus GAAUUGGGUGAGUUGAGGAUGAGCCAGGCCACCGACACCGAGCACUGGAGCACCAUCCAGGCCUUGUUCGGUGAAGGAGAGC__AAAGAAGUCAACUC________ .........(((((((((.....))......((..((((((((((((((......)))))....)))))))))..))..............)))))))........ (-17.06 = -19.45 + 2.39)

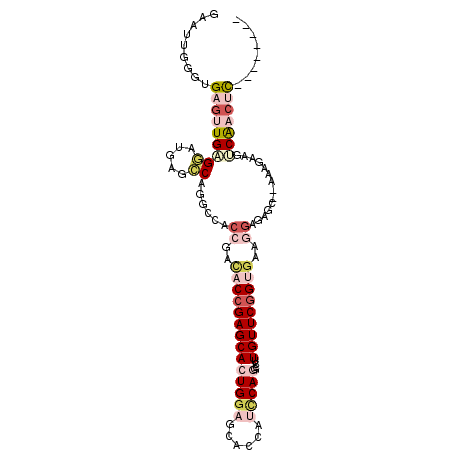
| Location | 831,563 – 831,659 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
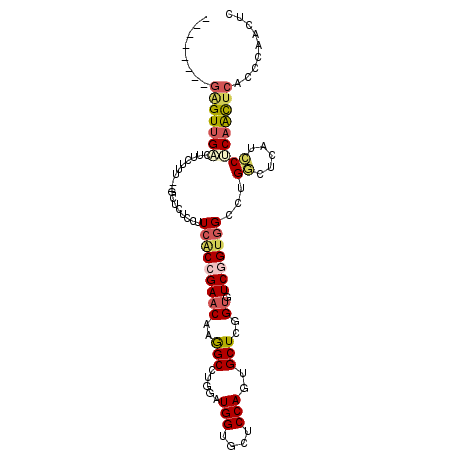
>3L_DroMel_CAF1 831563 96 - 23771897 --------GAGUUGACUUCUUG--GCUCUCCUUCACCGAACAAGGCCUGGAUGGUGCUCCAGUGCUCGGUGUCGGUGGCCUGGCUCAUCCUCAACUCACCCAAUUC --------(((((((......(--(((..((..((((((.....(((((((......))))).))))))))..)).)))).((.....)))))))))......... ( -35.10) >DroPse_CAF1 6770 105 - 1 GGCACGGGGAGUCGUCUCCCCAGUGCC-CUGUUCACCGAACAGAGCGUGUAUGGUCUGCCAGUGCUCCGAAUCUGUAGAUUGUCGCACCCUCAGCUCGGACAGCUC (((((((((((....)))))).)))))-((((...((((.(.(((.(((((..((((((.((..........))))))))..).)))).))).).))))))))... ( -45.30) >DroSec_CAF1 6051 96 - 1 --------GAGUUGACUUCUUU--GCUCUCCAUCGCCGAACAAGGCCUGGAUGGUGCUCCAGUGCUCGGUGUCGGUGGCCUGGCUCAUCCUCAACUCACCCAAUUC --------(((((((.......--(((.....(((((((((..((((((((......))))).)))..)).)))))))...)))......)))))))......... ( -33.32) >DroSim_CAF1 5479 95 - 1 --------GAGUUGACUCCUUU--GCU-UCCAUCACCGAACAAGGCCUGGAUGGUGCUCCAGUGCUCGGUGUCGGUGGCCUGGCUCAUCCUCAACUCACCCAAUUC --------(((((((.......--(((-....(((((((((..((((((((......))))).)))..)).)))))))...)))......)))))))......... ( -35.02) >DroEre_CAF1 6898 96 - 1 --------GGGUUGCCUUCUUG--GCCCUCCGUCACCGAACAAGGCUUGAAUGGUGCUCCAGUGCUCGGUGUCGGUGGCCUGGCUUAUCCGCAACUCUCCCAACUC --------(((((((......(--(((..(((.((((((.((.(((.........)))....)).)))))).))).)))).((.....)))))))))......... ( -37.10) >DroAna_CAF1 6486 96 - 1 --------AUGUUGAAUUUCAU--UAGAUCCUUCACCGAACAAGGCCUGAAUGGUCUGCCAGUGCUCGGUUUCUGAGGCCUGACGAAGUCGCAGUUCAUUUAACUC --------..(((((((....(--((((......(((((...(((((.....)))))((....))))))).)))))(..(((.((....)))))..)))))))).. ( -24.20) >consensus ________GAGUUGACUUCUUU__GCUCUCCUUCACCGAACAAGGCCUGGAUGGUGCUCCAGUGCUCGGUGUCGGUGGCCUGGCUCAUCCUCAACUCACCCAACUC ........(((((((.................(((((((((..(((.....(((....)))..)))..)).)))))))...((.....)))))))))......... (-17.12 = -17.85 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:32 2006