
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,366,792 – 8,366,951 |
| Length | 159 |
| Max. P | 0.901648 |

| Location | 8,366,792 – 8,366,911 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -36.41 |
| Energy contribution | -37.23 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8366792 119 - 23771897 ACUUUGGUUCCGUCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGAUUCCAGCGUGUCACCUACCACCUGGACUGGAUUCGCGACCAUACCGGCAUUGCUUACUACUAAUCGAC-UAUAUCU ...(((((...(((((.((...(((((((((((..(.((((((...)))))).).)))))..(....)))))))..)))))))...))))).....................-....... ( -37.20) >DroSec_CAF1 3213 119 - 1 ACUUUGGUUCCGUCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGCUUCCAGCGGGUCACCUACCACCUGGACUGGAUUCGCGACCAUACCGGCAUUGCUUACUACUAAACGAC-UAAAUCU ...(((((...(((((.((...(((((((((((..((((((((...)))))))).)))))..(....)))))))..)))))))...))))).....................-....... ( -44.30) >DroYak_CAF1 3786 119 - 1 ACUUCGGUUCCUCCGCCGGCUGCCAGUCGGGUGCUCCUGCUGGCUUCCAGCGUGUGACCUACCACCUGGACUGGAUCCGCGACCACACCGGCAUCGCUUACUACUAAACGAC-UAUAUCU .....((.((((((((.((..(((((.((((....)))))))))..)).))(.(((......)))).)))..))).))((((((.....))..))))...............-....... ( -37.70) >DroAna_CAF1 4926 114 - 1 ACUUCGGCUCCUCUGCUGGCUGCCAGUCGGGUGCUCCUGCUGGUUUCCAGCGGGUUACCUACCACCUGGACUGGAUCCGCGACCACACCGGUAUUGCUUACU---AAACGAAAUAAA--- ..(((((.(((((((((((..(((((.((((....)))))))))..))))))))...((........))...))).))((.(((.....)))...)).....---....))).....--- ( -40.20) >consensus ACUUCGGUUCCGCCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGCUUCCAGCGGGUCACCUACCACCUGGACUGGAUCCGCGACCACACCGGCAUUGCUUACUACUAAACGAC_UAAAUCU ...(((((...(((((.((...(((((((((((..((((((((...))))))))........))))).))))))..)))))))...)))))............................. (-36.41 = -37.23 + 0.81)



| Location | 8,366,831 – 8,366,951 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -53.85 |
| Consensus MFE | -42.11 |
| Energy contribution | -42.80 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8366831 120 + 23771897 GCGAAUCCAGUCCAGGUGGUAGGUGACACGCUGGAAUCCAGCGGGAGCACCCGACUGGCAGCCGGCGACGGAACCAAAGUUGGUGACCCCCACAAGCUUGGUACCGUCAUGGGUGACCAG ((....((((((..((((.(.(....).((((((...))))))..).)))).))))))..))....(((((.(((((.(((.(((.....))).)))))))).))))).(((....))). ( -53.90) >DroSec_CAF1 3252 120 + 1 GCGAAUCCAGUCCAGGUGGUAGGUGACCCGCUGGAAGCCAGCGGGAGCACCCGACUGGCAGCCGGCGACGGAACCAAAGUUAGUGACUCCCACAAGCUUGGUACCGUCAUGGGUGACCAG ......(((.......)))..(((.(((((((((..(((((((((....)))).)))))..)))))(((((.(((((.(((.(((.....))).)))))))).)))))..)))).))).. ( -61.10) >DroYak_CAF1 3825 120 + 1 GCGGAUCCAGUCCAGGUGGUAGGUCACACGCUGGAAGCCAGCAGGAGCACCCGACUGGCAGCCGGCGGAGGAACCGAAGUUGGUGACUCCCACAAGCUUGGUGCCGUCAUGGGUGACCAG ((((..((((.(...((((.........((((((..(((((..((.....))..)))))..))))))(((..((((....))))..)))))))..).))))..))))..(((....))). ( -52.60) >DroAna_CAF1 4960 120 + 1 GCGGAUCCAGUCCAGGUGGUAGGUAACCCGCUGGAAACCAGCAGGAGCACCCGACUGGCAGCCAGCAGAGGAGCCGAAGUUGGUGACACCUACCAGCUUGGUACCGUCGUGUGUGACCAG ((((..((((((..((((.(.((....))(((((...)))))...).)))).))))))...)).))...((.(((.(((((((((.....)))))))))))).))((((....))))... ( -47.80) >consensus GCGAAUCCAGUCCAGGUGGUAGGUGACACGCUGGAAGCCAGCAGGAGCACCCGACUGGCAGCCGGCGAAGGAACCAAAGUUGGUGACUCCCACAAGCUUGGUACCGUCAUGGGUGACCAG ......(((.......)))..(((.((.((((((..(((((((((....)))).)))))..)))))(((((.(((((.(((.(((.....))).)))))))).)))))..).)).))).. (-42.11 = -42.80 + 0.69)


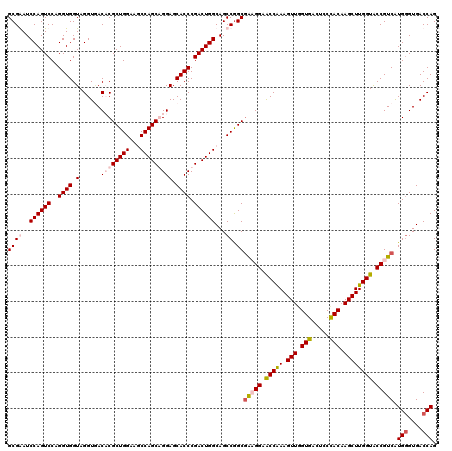
| Location | 8,366,831 – 8,366,951 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -49.62 |
| Consensus MFE | -41.06 |
| Energy contribution | -41.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8366831 120 - 23771897 CUGGUCACCCAUGACGGUACCAAGCUUGUGGGGGUCACCAACUUUGGUUCCGUCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGAUUCCAGCGUGUCACCUACCACCUGGACUGGAUUCGC .((((.(((......)))))))((((.((((.((..((((....)))).)).)))).)))).(((((((((((..(.((((((...)))))).).)))))..(....)))))))...... ( -47.30) >DroSec_CAF1 3252 120 - 1 CUGGUCACCCAUGACGGUACCAAGCUUGUGGGAGUCACUAACUUUGGUUCCGUCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGCUUCCAGCGGGUCACCUACCACCUGGACUGGAUUCGC ..(((.((((..(((((.((((((.(((((.....))).)).)))))).)))))((.((..(((((.((((....)))))))))..)).)))))).))).....((......))...... ( -53.50) >DroYak_CAF1 3825 120 - 1 CUGGUCACCCAUGACGGCACCAAGCUUGUGGGAGUCACCAACUUCGGUUCCUCCGCCGGCUGCCAGUCGGGUGCUCCUGCUGGCUUCCAGCGUGUGACCUACCACCUGGACUGGAUCCGC ...((((....))))((..(((..((.((((..(((((.(((....)))....(((.((..(((((.((((....)))))))))..)).))).)))))...))))..))..)))..)).. ( -47.30) >DroAna_CAF1 4960 120 - 1 CUGGUCACACACGACGGUACCAAGCUGGUAGGUGUCACCAACUUCGGCUCCUCUGCUGGCUGCCAGUCGGGUGCUCCUGCUGGUUUCCAGCGGGUUACCUACCACCUGGACUGGAUCCGC ..((((.(....).((((.(((.(.(((((((((...((..(....).......(((((..(((((.((((....)))))))))..)))))))..)))))))))).)))))))))))... ( -50.40) >consensus CUGGUCACCCAUGACGGUACCAAGCUUGUGGGAGUCACCAACUUCGGUUCCGCCGCCGGCUGCCAGUCGGGUGCUCCCGCUGGCUUCCAGCGGGUCACCUACCACCUGGACUGGAUCCGC ...(((......)))((..((((((..(((((((.((((....(((((.((......))..)))))...)))))))))))..)))(((((..(((.....)))..))))).)))..)).. (-41.06 = -41.25 + 0.19)

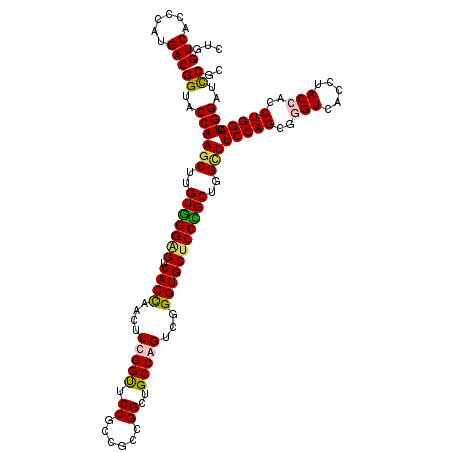

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:10 2006