
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,364,472 – 8,364,612 |
| Length | 140 |
| Max. P | 0.969684 |

| Location | 8,364,472 – 8,364,580 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -31.84 |
| Energy contribution | -32.34 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8364472 108 - 23771897 CCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGAAUCUGGGAAAACCCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAGGUCAUAA .......(((((((((.....((((....((((((((((.....((.....))))))))))..))..)))).)))))))))(((((...(((...)))...))))).. ( -35.60) >DroPse_CAF1 865 99 - 1 UCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGG-----GG----AGCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAAAUCAUAA ...(((.(((((((((.....((((....((((((((((.-----..----..))))))))..))..)))).)))))))))(((.(...).))).))).......... ( -31.30) >DroSec_CAF1 887 107 - 1 CCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGGAUCUGGGA-AACCCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAGGUCAUAA .......(((((((((.....((((....(((((((((((.....(..-..))))))))))..))..)))).)))))))))(((((...(((...)))...))))).. ( -38.60) >DroEre_CAF1 920 107 - 1 CCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGGAUCUGGAA-AACCCCCUUUGAUAGCAAGGUGGCAAUUAGGCGUGGCAAGGGCACAGCUAAGGUCAUAA .......((..(((((.....((((....(((((((((((........-...)))))))))..))..)))).)))))..))(((((...(((...)))...))))).. ( -32.70) >DroYak_CAF1 1273 107 - 1 CCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGCAUCGGGAU-GACCCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAGGUCAUAA .......(((((((((.....((((....((((((((((....((...-..))))))))))..))..)))).)))))))))(((((...(((...)))...))))).. ( -36.00) >DroPer_CAF1 865 99 - 1 UCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGG-----GG----AGCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAAAUCAUAA ...(((.(((((((((.....((((....((((((((((.-----..----..))))))))..))..)))).)))))))))(((.(...).))).))).......... ( -31.30) >consensus CCCAGUGGCUCAAUUGACAAACAUCACUGGCUCAAAGGGGAUCUGGGA_AACCCCCUUUGAUAGCAAGGUGGCAAUUGGGCGUGGCAAGGGCGCAGCUAAGGUCAUAA .......(((((((((.....((((....((((((((((..............))))))))..))..)))).)))))))))(((((...(((...)))...))))).. (-31.84 = -32.34 + 0.50)



| Location | 8,364,507 – 8,364,612 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -18.19 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8364507 105 + 23771897 GCCACCUUGCUAUCAAAGGGGGUUUUCCCAGAU----UCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA--GCCACUGGGCACAACCCCAAU--CCACAUAUGACACCACACGC (((.....((..((((((((((((......)))----)))))))))))(((((..(((((.....)))--))))))))))...........--.................... ( -28.20) >DroPse_CAF1 900 98 + 1 GCCACCUUGCUAUCAAAGGGCU----CC---------CCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA--GCCACUGGACGCAUCCCUAAAAGUCCCAUAUCCCCCCUGCCCC .......(((..((((((((..----..---------.))))))))(((((((..(((((.....)))--)))))))).)))).............................. ( -23.50) >DroSec_CAF1 922 104 + 1 GCCACCUUGCUAUCAAAGGGGGUU-UCCCAGAU----CCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA--GCCACUGGGCACAACCCCAAU--CCACAUAUGACACCACACGC (((.....((..((((((((((((-.....)))----)))))))))))(((((..(((((.....)))--))))))))))...........--.................... ( -30.80) >DroYak_CAF1 1308 104 + 1 GCCACCUUGCUAUCAAAGGGGGUC-AUCCCGAU----GCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA--GCCACUGGGCACAACCCUAAU--CCACAUCUAACACCACACGC (((.....((..((((((((.(((-.....)))----.))))))))))(((((..(((((.....)))--))))))))))...........--.................... ( -29.00) >DroAna_CAF1 906 103 + 1 GCCGCCUUGCUAUCAAAGGCGGUCCAACCCGUUGAGCCCCCUUUGAGCCAGUGAUGUUUGUCAAUUGAGUGCCACUGGGCGCAUCCCUAAG--CCACAU------CCACAC-- ........((..(((((((.((..(((....)))..)).)))))))))..(((((((..((.....(((((((....))))).)).....)--).))))------.)))..-- ( -29.90) >DroPer_CAF1 900 98 + 1 GCCACCUUGCUAUCAAAGGGCU----CC---------CCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA--GCCACUGGACGCAUCCCUAAAAGUCCCAUAUCCCCCCUGCCCC .......(((..((((((((..----..---------.))))))))(((((((..(((((.....)))--)))))))).)))).............................. ( -23.50) >consensus GCCACCUUGCUAUCAAAGGGGGU___CCC_GAU____CCCCUUUGAGCCAGUGAUGUUUGUCAAUUGA__GCCACUGGGCACAACCCUAAU__CCACAUAUCACACCACACGC .......(((..((((((((..................)))))))).((((((..((...((....))..)))))))))))................................ (-18.19 = -18.44 + 0.25)

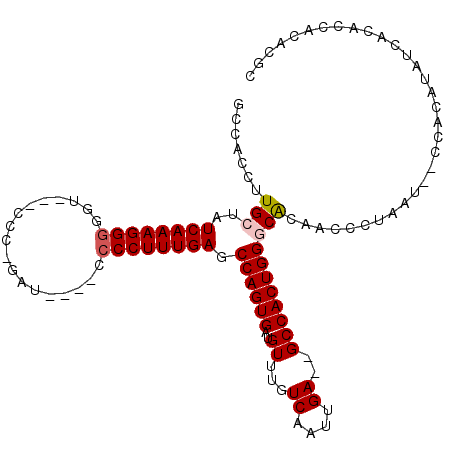
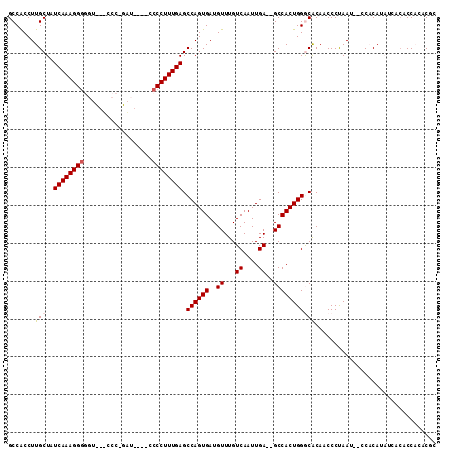
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:06 2006