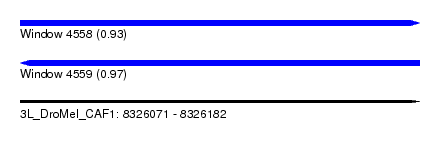
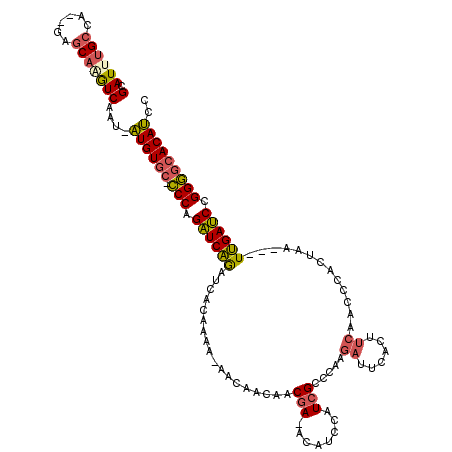
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,326,071 – 8,326,182 |
| Length | 111 |
| Max. P | 0.970313 |

| Location | 8,326,071 – 8,326,182 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -29.53 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8326071 111 + 23771897 GGAUGUGCCCCGGAUCAA---UUAGUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU-UCGUUGUUGUU-UUUUGUGAUCUGAUCUGGG-GCACAU-AUUGAUCUGCUC--UGGCAAAUGC ..((((((((((((((((---(((..(((((......)))))..((((((((....-))))))))...-.....)))).)))))))))-))))))-.......(((..--..)))..... ( -37.70) >DroSec_CAF1 4914 114 + 1 GGAUGUGCCCCGGAUCAA---UUAUUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU-UCGUUGUUGUU-UUUUGUGAUCUGAUCUGGG-GCACAUGACUGACUUGCUCUUUGGCAAAUGC ..((((((((((((((((---((((.(((((......)))))..((((((((....-))))))))...-....))))).)))))))))-)))))).......(((((....))))).... ( -39.60) >DroSim_CAF1 4907 114 + 1 GGAUGUGCCCCGGAUCAA---UUAGUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU-UCGUUGUUGUU-UUUUGUGAUCUGAUCUGGG-GCACAUGACUGACUUGCUCUUUGGCAAAUGC ..((((((((((((((((---(((..(((((......)))))..((((((((....-))))))))...-.....)))).)))))))))-)))))).......(((((....))))).... ( -38.90) >DroEre_CAF1 4965 108 + 1 GGAUGUGCCCCAGAUCAA---UUAGUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU-UCGUUGUUA----UUUGUGAUCUGAUCUGGG-GCACAU-AUUGAUUUGCUA--UGGCAAAUGC ..((((((((((((((((---(((..(((((......)))))..((((((((....-)))))))).----....)))).)))))))))-))))))-....((((((..--..)))))).. ( -41.50) >DroYak_CAF1 5004 108 + 1 GGAUGUGCCCCAGAUCAA---UUAGUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU-UCGUUGUUA----UUUGUGAUCUGAUCUGGG-GCACAU-AUUGACUUGCUC--UGGCACAUGC ..((((((((((((((((---(((..(((((......)))))..((((((((....-)))))))).----....)))).)))))))))-))))))-.......(((..--..)))..... ( -38.20) >DroAna_CAF1 5740 116 + 1 AAUUGUUAUCCCGGUCGAGAUUCAAGGGAUUGGAAUCA-UUUGAGGCUAAGAGCGUAUUGUUGUUGACUGUGUGUGACUCGACCUGGACGCACAA-UUUGACAUGACC--UGGCUAAUGC ((((((..(((.(((((((.(((((....))))).(((-(....((.(((.(((.....))).))).))....))))))))))).)))...))))-))..........--.......... ( -28.30) >consensus GGAUGUGCCCCGGAUCAA___UUAGUGGGUUGAAGUGAAUCUUGGGCGAUGGAUGU_UCGUUGUUGUU_UUUUGUGAUCUGAUCUGGG_GCACAU_AUUGACUUGCUC__UGGCAAAUGC ..(((((((((((((((.........(((((......)))))..((((((((.....))))))))..............))))))))).))))))........((((....))))..... (-29.53 = -28.90 + -0.63)

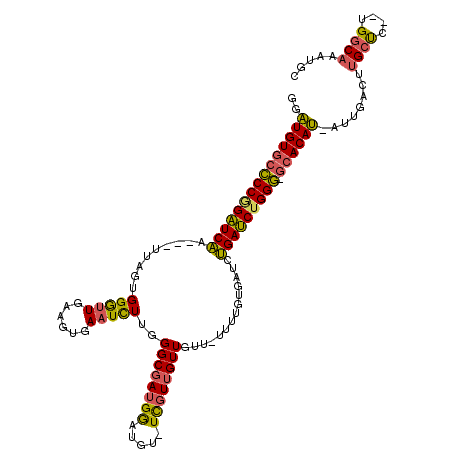
| Location | 8,326,071 – 8,326,182 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -21.02 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8326071 111 - 23771897 GCAUUUGCCA--GAGCAGAUCAAU-AUGUGC-CCCAGAUCAGAUCACAAAA-AACAACAACGA-ACAUCCAUCGCCCAAGAUUCACUUCAACCCACUAA---UUGAUCCGGGGCACAUCC (.((((((..--..)))))))...-((((((-(((.((((((.........-........(((-.......))).....((......))..........---)))))).))))))))).. ( -29.10) >DroSec_CAF1 4914 114 - 1 GCAUUUGCCAAAGAGCAAGUCAGUCAUGUGC-CCCAGAUCAGAUCACAAAA-AACAACAACGA-ACAUCCAUCGCCCAAGAUUCACUUCAACCCAAUAA---UUGAUCCGGGGCACAUCC (.((((((......)))))))....((((((-(((.((((((.........-........(((-.......))).....((......))..........---)))))).))))))))).. ( -29.30) >DroSim_CAF1 4907 114 - 1 GCAUUUGCCAAAGAGCAAGUCAGUCAUGUGC-CCCAGAUCAGAUCACAAAA-AACAACAACGA-ACAUCCAUCGCCCAAGAUUCACUUCAACCCACUAA---UUGAUCCGGGGCACAUCC (.((((((......)))))))....((((((-(((.((((((.........-........(((-.......))).....((......))..........---)))))).))))))))).. ( -29.30) >DroEre_CAF1 4965 108 - 1 GCAUUUGCCA--UAGCAAAUCAAU-AUGUGC-CCCAGAUCAGAUCACAAA----UAACAACGA-ACAUCCAUCGCCCAAGAUUCACUUCAACCCACUAA---UUGAUCUGGGGCACAUCC (.((((((..--..)))))))...-((((((-((((((((((........----......(((-.......))).....((......))..........---)))))))))))))))).. ( -33.60) >DroYak_CAF1 5004 108 - 1 GCAUGUGCCA--GAGCAAGUCAAU-AUGUGC-CCCAGAUCAGAUCACAAA----UAACAACGA-ACAUCCAUCGCCCAAGAUUCACUUCAACCCACUAA---UUGAUCUGGGGCACAUCC .....(((..--..))).......-((((((-((((((((((........----......(((-.......))).....((......))..........---)))))))))))))))).. ( -30.90) >DroAna_CAF1 5740 116 - 1 GCAUUAGCCA--GGUCAUGUCAAA-UUGUGCGUCCAGGUCGAGUCACACACAGUCAACAACAAUACGCUCUUAGCCUCAAA-UGAUUCCAAUCCCUUGAAUCUCGACCGGGAUAACAAUU ((....))..--..........((-((((..((((.(((((((.......................((.....)).((((.-.(((....)))..))))..))))))).)))).)))))) ( -25.30) >consensus GCAUUUGCCA__GAGCAAGUCAAU_AUGUGC_CCCAGAUCAGAUCACAAAA_AACAACAACGA_ACAUCCAUCGCCCAAGAUUCACUUCAACCCACUAA___UUGAUCCGGGGCACAUCC (.((((((......)))))))....((((((.(((.((((((..................(((........))).....((......)).............)))))).))))))))).. (-21.02 = -20.97 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:03 2006