
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,307,779 – 8,307,965 |
| Length | 186 |
| Max. P | 0.872776 |

| Location | 8,307,779 – 8,307,869 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -12.88 |
| Energy contribution | -11.62 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
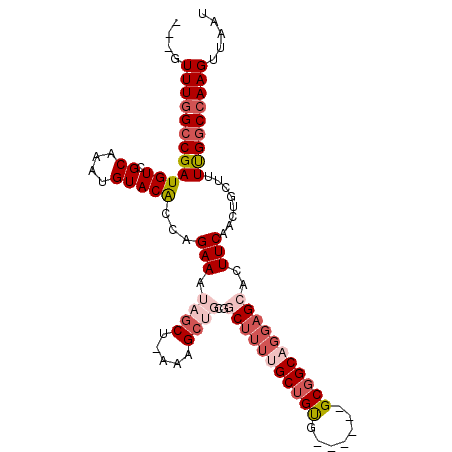
>3L_DroMel_CAF1 8307779 90 + 23771897 GGUUCCUUAAUACAUUAACAAACAGAGAAAUGUUUCUUGAUAUUUUUUGCAGUGUGACAAACUUUUUGUCCGAUUGCUUUGUUUUAUUUU ................((((((..((((((((((....))))))))))(((((..(((((.....)))))..)))))))))))....... ( -16.80) >DroSec_CAF1 14095 90 + 1 AGUUAAUUAAUACAUGAAUAAACAGAGCAAUGCUGCUUGAUAUUUUUUGCAGUGUGACAAACUUUUUGUCCGAUUGCUUUGUUUUAUUUU ...............(((((((((((((((((((((..((....))..)))))(.(((((.....)))))).))))))))).))))))). ( -24.70) >DroSim_CAF1 12734 90 + 1 AGUUAAUUAAUACAUUAAUAAACAGAGCAAUGUUGCUUGGUAUUUUUUGCAGUGUGACAAACUUUUUGUCCGAUUGCUUUGUUUUAUUUU .((((((......))))))((((((((((((.((((..((....))..))))((.(((((.....)))))))))))))))))))...... ( -20.00) >DroEre_CAF1 13326 86 + 1 AGUUUCUUUAUAAAUUAAUAAACAGAAAAAGGUUUU---CCUCUUUUUGCAGUGUGGCAAACUU-UUGUCCAAUUGUUUUGUUUUAUUUU ...........((((.((((((((((((.(((....---))).))))))((((.((((((....-))).))))))).))))))..)))). ( -11.70) >consensus AGUUAAUUAAUACAUUAAUAAACAGAGAAAUGUUGCUUGAUAUUUUUUGCAGUGUGACAAACUUUUUGUCCGAUUGCUUUGUUUUAUUUU ...................((((((((((....)))............(((((..(((((.....)))))..))))))))))))...... (-12.88 = -11.62 + -1.25)



| Location | 8,307,869 – 8,307,965 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -22.47 |
| Energy contribution | -24.55 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8307869 96 + 23771897 ---GUUUGGCCGAUGUCGCAAAUGUACACCAGAAAUAGCU-AUAGCUUCGGCUUUUGCUGUA------GCGGCAGGAGGACUUCAACUGCUUUUGGCCAAGUUAAU ---.(((((((((((((((...((.....))...(((((.-..(((....)))...))))).------))))))(.((........)).)..)))))))))..... ( -33.30) >DroSec_CAF1 14185 96 + 1 ---GUUUGGCCGAUGUCGCAAAUGUACACAAGAAAUAGCU-AGAGCUGCGGCUUUUGCUGUG------GCGGCAGGAGCACUUCAACUGCUUUUGGCCAAGUUAAU ---.(((((((((((((((...((....))....(((((.-(((((....))))).))))).------)))))).(((((.......))))))))))))))..... ( -35.00) >DroSim_CAF1 12824 96 + 1 ---GUUUGGCCGAUGUCGCAAAUGUACACCAGAAAUAGCU-AGAGCUGCCGCUUUUGCUGUG------GCGGCAGGAGCACUUCAACUGCUUUUGGCCAAGUUAAU ---.(((((((((((((((...((.....))...(((((.-(((((....))))).))))).------)))))).(((((.......))))))))))))))..... ( -33.70) >DroEre_CAF1 13412 102 + 1 ---GUUUGGCCGAUGUCGCAAAUGUACACCAGAAAUAGCU-AUAGCAGUGGCUUUUGCUGUGGCCGCGGCGGCAGGAGCACUUCAACUGCUUUUGGCCAAGUUAAU ---.(((((((((.(((((...((.....))......(((-(((((((......)))))))))).)))))(((((...........))))).)))))))))..... ( -38.50) >DroYak_CAF1 13384 105 + 1 UUUUUUUCGCCGAUGUCGCAAAUGUACACCAGAAAUAGCU-AAAGCUGCGGCUUUGGCUGUGGCCGCGGCGGCAGGAGCACUUCAACUGCUUUUGGCCAAGUUAAU ........((((..(((((...((.....)).....((((-(((((....))))))))))))))..))))((((((((((.......))))))).)))........ ( -37.00) >DroPer_CAF1 15065 82 + 1 ---GUUUGGACGAUGUCGCAAAUGUACGGCAGAAACCUCAGAAAG------C---AGCAGCA------GCAGCAGGAGCAGUUCAG------UCGGCCAAGUUAAU ---.(((((.((((((((........)))).(((..(((.....(------(---.((....------)).))..)))...))).)------))).)))))..... ( -19.30) >consensus ___GUUUGGCCGAUGUCGCAAAUGUACACCAGAAAUAGCU_AAAGCUGCGGCUUUUGCUGUG______GCGGCAGGAGCACUUCAACUGCUUUUGGCCAAGUUAAU ....((((((((((((.((....)))))...(((.((((.....))))..(((((((((((.......)))))))))))..)))........)))))))))..... (-22.47 = -24.55 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:57 2006