
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,293,827 – 8,293,966 |
| Length | 139 |
| Max. P | 0.996478 |

| Location | 8,293,827 – 8,293,939 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -24.87 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293827 112 + 23771897 AUGAUAUAUGCAUUUUUCAAGCAUCGCAUCGCAUCGCCAACAAAGACUGCCUC--------UUGCUUAUAAAUUAGAAGCAACCGAAAUAAUAGCCAUAUUUUGGUUGCUUUUCGCCAGU .........((.......(((((..(((((..............)).)))...--------.))))).......((((((((((((((((.......)))))))))))))))).)).... ( -26.04) >DroSim_CAF1 15313 112 + 1 AUGAUAUAUGCAUUUUUCAAGCAUCGCAUCGCAUCGCCAACAAAGGCUGCCUC--------UUGCUUAUAAAUUAGAAGCAACCGAAAUAAUAGCCAUAUUUUGGUUGCUUUUCGCCAGU .........((.......(((((.......(((..(((......))))))...--------.))))).......((((((((((((((((.......)))))))))))))))).)).... ( -29.40) >DroEre_CAF1 15804 110 + 1 AUGAUAUAUGCAUUUUUCAAGCAU----------CGCCAACAAUGGCUGCCUCUGCGCGCUUUGCUUGUAAAUUGGAAGCAGCCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGU ..........((.(((.((((((.----------.((((....)))).((......))....)))))).))).))(((((((((((((((.......)))))))))))))))........ ( -32.50) >DroYak_CAF1 15341 110 + 1 AUGAUAUAUGCAUUUUUCAAGCAU----------CGCCAACAAUGGCUGCCUCUGCGCGCGUUGCUUAUAAAUUAGAAGCAGCCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGU .........((.......(((((.----------(((((....))((.((....))))))).))))).......((((((((((((((((.......)))))))))))))))).)).... ( -30.50) >consensus AUGAUAUAUGCAUUUUUCAAGCAU__________CGCCAACAAAGGCUGCCUC________UUGCUUAUAAAUUAGAAGCAACCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGU .........((.......(((((............(((......)))...............))))).......((((((((((((((((.......)))))))))))))))).)).... (-24.87 = -24.68 + -0.19)

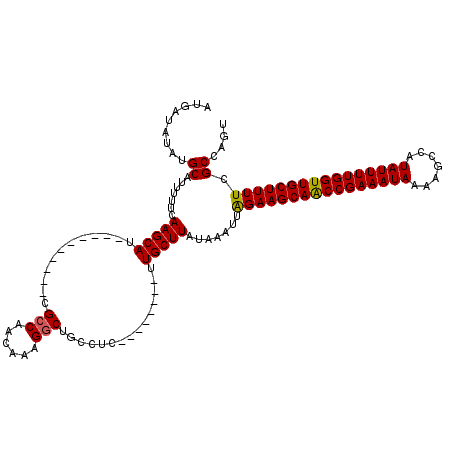
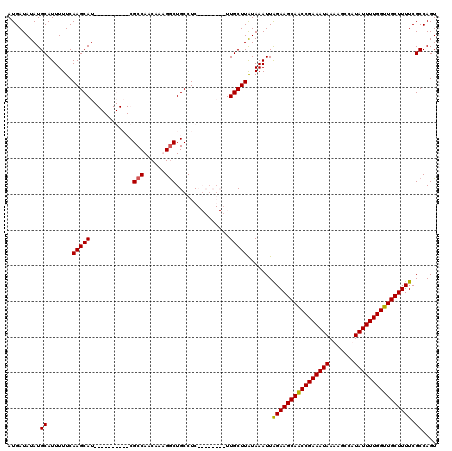
| Location | 8,293,867 – 8,293,966 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -20.58 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293867 99 + 23771897 CAAAGACUGCCUC--------UUGCUUAUAAAUUAGAAGCAACCGAAAUAAUAGCCAUAUUUUGGUUGCUUUUCGCCAGUUUUGGUUUUU-------------GCUUUUCCAACACAUAU (((((((((...(--------(.((.........((((((((((((((((.......)))))))))))))))).)).))...))))))))-------------)................ ( -24.50) >DroSim_CAF1 15353 97 + 1 CAAAGGCUGCCUC--------UUGCUUAUAAAUUAGAAGCAACCGAAAUAAUAGCCAUAUUUUGGUUGCUUUUCGCCAGUUUUGGUUUUU-------------GCUUUUCCAACAUAU-- .((((((.(((..--------..(((........((((((((((((((((.......))))))))))))))))....)))...)))....-------------)))))).........-- ( -25.60) >DroEre_CAF1 15834 120 + 1 CAAUGGCUGCCUCUGCGCGCUUUGCUUGUAAAUUGGAAGCAGCCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGUUUUGGUUUUUGGUUUUGGUUUUCGGUUUUCCAGCUCUUUU ....((((((....))).)))..(((.(.(((((((((...((((((((((((((((....(((((........)))))...)))))))).))))))))))))))))).).)))...... ( -33.10) >DroYak_CAF1 15371 108 + 1 CAAUGGCUGCCUCUGCGCGCGUUGCUUAUAAAUUAGAAGCAGCCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGUUUUGGUUU------------UUCGAUUUACCAUCUUUUUU (((((((.((....)))).)))))....((((((((((((((((((((((.......)))))))))))))))).(((......)))..------------...))))))........... ( -28.00) >consensus CAAAGGCUGCCUC________UUGCUUAUAAAUUAGAAGCAACCGAAAUAAAAGCCAUAUUUUGGUUGCUUUUCGCCAGUUUUGGUUUUU_____________GCUUUUCCAACACAUUU (((.(((((.(..........(((....)))...((((((((((((((((.......)))))))))))))))).).)))))))).................................... (-20.58 = -19.95 + -0.63)

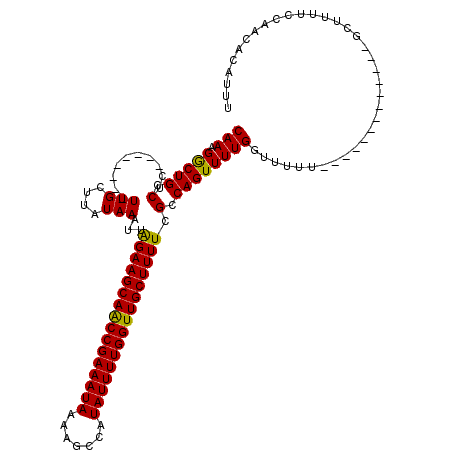

| Location | 8,293,867 – 8,293,966 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293867 99 - 23771897 AUAUGUGUUGGAAAAGC-------------AAAAACCAAAACUGGCGAAAAGCAACCAAAAUAUGGCUAUUAUUUCGGUUGCUUCUAAUUUAUAAGCAA--------GAGGCAGUCUUUG .((((.(((((((....-------------.....(((....)))......((((((.(((((.......))))).))))))))))))).)))).((..--------...))........ ( -20.70) >DroSim_CAF1 15353 97 - 1 --AUAUGUUGGAAAAGC-------------AAAAACCAAAACUGGCGAAAAGCAACCAAAAUAUGGCUAUUAUUUCGGUUGCUUCUAAUUUAUAAGCAA--------GAGGCAGCCUUUG --...((((.....)))-------------)............(((...((((((((.(((((.......))))).))))))))...........((..--------...)).))).... ( -21.00) >DroEre_CAF1 15834 120 - 1 AAAAGAGCUGGAAAACCGAAAACCAAAACCAAAAACCAAAACUGGCGAAAAGCAACCAAAAUAUGGCUUUUAUUUCGGCUGCUUCCAAUUUACAAGCAAAGCGCGCAGAGGCAGCCAUUG ..(((((((((....))...........................((.....))...........))))))).....(((((((((..........((.....))...))))))))).... ( -24.92) >DroYak_CAF1 15371 108 - 1 AAAAAAGAUGGUAAAUCGAA------------AAACCAAAACUGGCGAAAAGCAACCAAAAUAUGGCUUUUAUUUCGGCUGCUUCUAAUUUAUAAGCAACGCGCGCAGAGGCAGCCAUUG ......(((.....)))(((------------(..(((....)))..((((((............)))))).))))((((((((((.........((.....))..)))))))))).... ( -25.70) >consensus AAAAAAGUUGGAAAAGC_____________AAAAACCAAAACUGGCGAAAAGCAACCAAAAUAUGGCUAUUAUUUCGGCUGCUUCUAAUUUAUAAGCAA________GAGGCAGCCAUUG ..........((((.....................(((....)))..((((((............)))))).))))(((((((((......................))))))))).... (-16.89 = -17.20 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:51 2006