| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 830,682 – 830,798 |
| Length | 116 |
| Max. P | 0.770736 |

| Location | 830,682 – 830,798 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
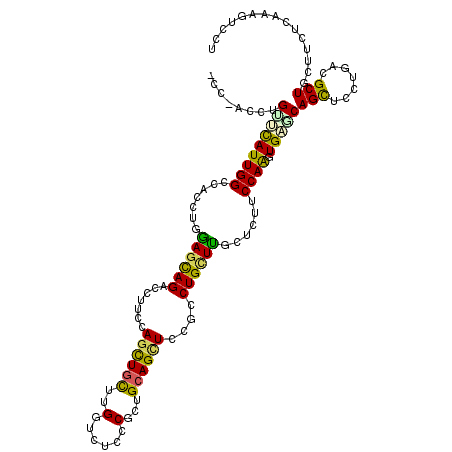
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -19.97 |
| Energy contribution | -21.58 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
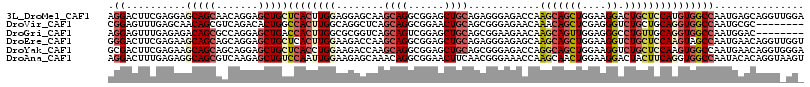
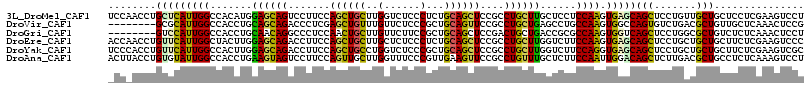
>3L_DroMel_CAF1 830682 116 + 23771897 AGGACUUCGAGGAGCAGCAACAGGAGCUGCUCACUUGGAGGAGCAAGCAGGCGGAGCUGCAGAGGGAGACCAAGCAGCUGGAAGGACUGCUCCAUGUGGCCAAUGAGCAGGUUGGA .(((((((((((((((((.......))))))).)))))))......((((.(..((((((...((....))..))))))....)..)))))))......(((((......))))). ( -48.60) >DroVir_CAF1 6523 108 + 1 CGGAGUUUGAGCAACAGCGUCAGACACUGGCCACUUGGCAGGCUCAGCAGGCGGAACUGCAGCGGGAGAACAAACAGCUCGAGGGUCUGCUGCAGGUGGCCAAUGCGC-------- ....((((((((....)).))))))..((((((((((((((((((.((((......))))..((((...........)))).))))))))..))))))))))......-------- ( -47.90) >DroGri_CAF1 4294 108 + 1 AGGAGUUUGAGAGACAGCGCCAGGAGCUGACCACUUGGCGCGGUCAGCAGUCGGAGCUGCAGCGGAAGAACAAGCAGUUGGAGGGCCUGUUGCAGGUGGCCAAUGGAC-------- ....(((((...(((.((((((((.........)))))))).))).(((((....)))))..(....)..))))).(((((...(((((...)))))..)))))....-------- ( -40.80) >DroEre_CAF1 6024 116 + 1 GGGACUUCGAGAAGCAGCAGCAGGAGCUGCUCACUUGGAAGACCAAGCAGGCGGAGCUGCAGAGGGAGAGCAAGCAGCUGGAAGGUCUGCUCCAAGUAGCCAAUGAACAGGUUGGU .............(.((((((....)))))))((((((((((((..((((......))))........(((.....)))....)))))..))))))).((((((......)))))) ( -38.90) >DroYak_CAF1 5485 116 + 1 GCGACUUCGAGAAGCAGCAGCAGGAGCUGCUCACCUGGAAGACCAAGCAGGCGGAGCUGCAGCGGGAGACCAGGCAGCUGGAAGGUCUGCUCCAAGUGGCCAAUGAACAGGUGGGA ....((((.((..(.((((((....)))))))..)).)))).((.(((((((..((((((...((....))..)))))).....))))))).......(((........))))).. ( -45.30) >DroAna_CAF1 5607 116 + 1 AGGACUUUGAGAGGCAGCGUCAAGAGCUGUCCAAUUGGAAGAGCAAACAGGCGGAACUUCAACGGGAAACCAAGCAACUGGAAGGACUACUUCAGGUGGCCAAUACACAGGUAAGU ((..((((.((..((..((((....(((.(((....)))..))).....))))..........((....))..))..)).))))..))(((((..(((.......)))..).)))) ( -32.60) >consensus AGGACUUCGAGAAGCAGCAGCAGGAGCUGCCCACUUGGAAGACCAAGCAGGCGGAGCUGCAGCGGGAGACCAAGCAGCUGGAAGGUCUGCUCCAGGUGGCCAAUGAACAGGU_GG_ .((..........(((((.......)))))((((((((........((((......))))............(((((((....)).)))))))))))))))............... (-19.97 = -21.58 + 1.61)
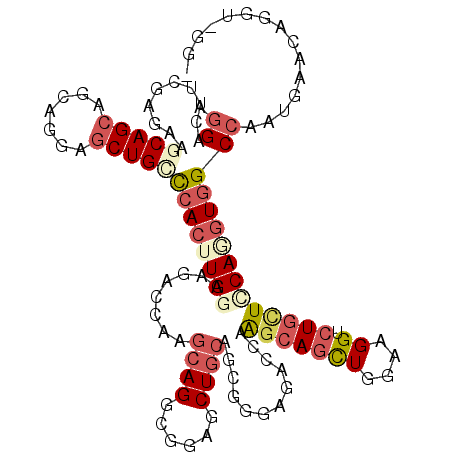
| Location | 830,682 – 830,798 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -19.10 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 830682 116 - 23771897 UCCAACCUGCUCAUUGGCCACAUGGAGCAGUCCUUCCAGCUGCUUGGUCUCCCUCUGCAGCUCCGCCUGCUUGCUCCUCCAAGUGAGCAGCUCCUGUUGCUGCUCCUCGAAGUCCU ......((((((..((....))..))))))..((((.((((((..((....))...))))))......(((((......)))))(((((((.......)))))))...)))).... ( -39.00) >DroVir_CAF1 6523 108 - 1 --------GCGCAUUGGCCACCUGCAGCAGACCCUCGAGCUGUUUGUUCUCCCGCUGCAGUUCCGCCUGCUGAGCCUGCCAAGUGGCCAGUGUCUGACGCUGUUGCUCAAACUCCG --------..(((((((((((.((..((((......((((.....))))....(((.((((.......))))))))))))).))))))))))).(((.((....)))))....... ( -38.70) >DroGri_CAF1 4294 108 - 1 --------GUCCAUUGGCCACCUGCAACAGGCCCUCCAACUGCUUGUUCUUCCGCUGCAGCUCCGACUGCUGACCGCGCCAAGUGGUCAGCUCCUGGCGCUGUCUCUCAAACUCCU --------.......(((.....(.(((((((.........))))))))....)))(((((.(((...(((((((((.....)))))))))...))).)))))............. ( -36.60) >DroEre_CAF1 6024 116 - 1 ACCAACCUGUUCAUUGGCUACUUGGAGCAGACCUUCCAGCUGCUUGCUCUCCCUCUGCAGCUCCGCCUGCUUGGUCUUCCAAGUGAGCAGCUCCUGCUGCUGCUUCUCGAAGUCCC ...............((((.(..(((((((.......(((((((..(.........((((......))))((((....)))))..))))))).......)))))))..).)))).. ( -37.94) >DroYak_CAF1 5485 116 - 1 UCCCACCUGUUCAUUGGCCACUUGGAGCAGACCUUCCAGCUGCCUGGUCUCCCGCUGCAGCUCCGCCUGCUUGGUCUUCCAGGUGAGCAGCUCCUGCUGCUGCUUCUCGAAGUCGC ...((((((......(((((.....(((((.(.....((((((.(((....)))..))))))..).))))))))))...))))))((((((....))))))((((....))))... ( -40.50) >DroAna_CAF1 5607 116 - 1 ACUUACCUGUGUAUUGGCCACCUGAAGUAGUCCUUCCAGUUGCUUGGUUUCCCGUUGAAGUUCCGCCUGUUUGCUCUUCCAAUUGGACAGCUCUUGACGCUGCCUCUCAAAGUCCU ......((.((....(((..(.(.(((.(((...((((((((...((....))...((((....((......)).))))))))))))..)))))).).)..)))...)).)).... ( -21.10) >consensus _CC_ACCUGUUCAUUGGCCACCUGGAGCAGACCUUCCAGCUGCUUGGUCUCCCGCUGCAGCUCCGCCUGCUUGCUCUUCCAAGUGAGCAGCUCCUGACGCUGCUUCUCAAAGUCCU ........(((((((((.......((((((.......((((((..(......)...))))))....))))))......)))).)))))(((.......)))............... (-19.10 = -18.30 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:28 2006