
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,293,004 – 8,293,187 |
| Length | 183 |
| Max. P | 0.776574 |

| Location | 8,293,004 – 8,293,107 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293004 103 + 23771897 GCUGACCA-----------------GAUCGAGAUCUCAGUUCCCCGUCUCCAGCUGCUCCUCCAUUUGCAGACGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUUCCAGCUUG ((((...(-----------------(((.(.((.......)).).)))).)))).((.....((((((((.....))))))))...((((((((...((....))..))))))))))... ( -28.40) >DroSec_CAF1 18932 103 + 1 GCUGACCA-----------------GAUCGAGAUCUCGGUCCACCGUCUCCAGCUGCUCCUCCAUUUGGAGUCGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUUCCAGCUUG ((((((((-----------------(((....)))).))))(((((.((((((.((......)).)))))).)))))..........(((((((...((....))..))))))))))... ( -37.90) >DroSim_CAF1 14466 103 + 1 GCUGACCA-----------------GAUCGAGAUCUCGGUCCACCGUCUCCAGCUGCUCCUCCAUUUGGAGUCGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUUCCAGCUUG ((((((((-----------------(((....)))).))))(((((.((((((.((......)).)))))).)))))..........(((((((...((....))..))))))))))... ( -37.90) >DroEre_CAF1 14806 117 + 1 GCUGACCAGGCCGAGACUCGUUCCAGAUCGAGAUCUCAGUCC-CCGUCUCCAGCUGCUCCUCCAUUUGGAGUUGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUU-CAGCUU- (((((...(((.((((((((........)))).)))).)))(-((((...((..(((.(((((....)))...)).)))..))...(((.........)))..)))))...)-))))..- ( -37.50) >DroYak_CAF1 14326 117 + 1 GCUGACCAGACCGAGACUCGUUCCAGAUCGAGAUCUCGUUGC-CCGUCUCCAGCUGCUCCUCCAUUUGGAGUUGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUU-CAGCUA- ((((((((...(((((((((........)))).)))))((((-(((.((((((.((......)).)))))).))).))))......)))......(((((......))))))-))))..- ( -40.70) >consensus GCUGACCA_________________GAUCGAGAUCUCGGUCC_CCGUCUCCAGCUGCUCCUCCAUUUGGAGUCGGUGCAAAUGAAAUGGAAAACGAAGCCAAAGCGGGUUUUCCAGCUUG ((((.(((.................((((((....))))))..(((.((((((.((......)).)))))).)))...........))).....((((((......)))))).))))... (-26.00 = -26.64 + 0.64)


| Location | 8,293,004 – 8,293,107 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -27.53 |
| Energy contribution | -27.29 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293004 103 - 23771897 CAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCGUCUGCAAAUGGAGGAGCAGCUGGAGACGGGGAACUGAGAUCUCGAUC-----------------UGGUCAGC ...((((.....(((((((..(((..((..(((..((((((((((.....))))))))))))))))))..))).))))..(((.((((....)))-----------------)))))))) ( -37.80) >DroSec_CAF1 18932 103 - 1 CAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCGACUCCAAAUGGAGGAGCAGCUGGAGACGGUGGACCGAGAUCUCGAUC-----------------UGGUCAGC ...((((((((((..((....))...)))))))..........(((((.(((((..((......))..))))).)))))((((.((((....)))-----------------)))))))) ( -37.20) >DroSim_CAF1 14466 103 - 1 CAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCGACUCCAAAUGGAGGAGCAGCUGGAGACGGUGGACCGAGAUCUCGAUC-----------------UGGUCAGC ...((((((((((..((....))...)))))))..........(((((.(((((..((......))..))))).)))))((((.((((....)))-----------------)))))))) ( -37.20) >DroEre_CAF1 14806 117 - 1 -AAGCUG-AAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCAACUCCAAAUGGAGGAGCAGCUGGAGACGG-GGACUGAGAUCUCGAUCUGGAACGAGUCUCGGCCUGGUCAGC -..((((-(...(((((((..(((.(.((((((((((...(((...)))....))))))))))).)))..))).)))-)..((((((.((((........)))))))))).....))))) ( -44.50) >DroYak_CAF1 14326 117 - 1 -UAGCUG-AAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCAACUCCAAAUGGAGGAGCAGCUGGAGACGG-GCAACGAGAUCUCGAUCUGGAACGAGUCUCGGUCUGGUCAGC -..((((-(...(((((((..(((.(.((((((((((...(((...)))....))))))))))).)))..))).)))-)...(((((.((((........)))))))))......))))) ( -43.40) >consensus CAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAUUUGCACCGACUCCAAAUGGAGGAGCAGCUGGAGACGG_GGACCGAGAUCUCGAUC_________________UGGUCAGC ...((((.....(((((((..(((.(.((((((((((................))))))))))).)))..))).))).)...(((....)))........................)))) (-27.53 = -27.29 + -0.24)



| Location | 8,293,067 – 8,293,187 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8293067 120 - 23771897 AAAAUUUCGCCCAGUGCACAUGUGUGCUUUUCUAUGGUCGCUUAUGUAAGGCAACCACCGUGCCGGCUUCUGCCCCAUUUCAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU ........(.(((..((((....)))).......))).)((((......((((.......))))(((....))).......))))((((((((..((....))...))))))))...... ( -30.40) >DroSec_CAF1 18995 120 - 1 ACAAUUUCGCCCAGUGCACAUGUGUGCUUUUCUAUGGUCGCUUAUGUAAGGCAACCACUGUGCCGGCUUCUGCCCCAUUUCAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU ........((.((((((((....))))........(((.((((.....)))).))))))).)).(((....)))...........((((((((..((....))...))))))))...... ( -32.90) >DroSim_CAF1 14529 120 - 1 ACAAUUUCGCCCAGUGUACAUGUGUGCUUUUCUAUGGUCGCUUAUGUAAGGCAACCACUGUGCCGGCUUCUGCCCCAUUUCAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU ........((.(((((((((((.(((((.......)).))).)))))).(....)))))).)).(((....)))...........((((((((..((....))...))))))))...... ( -31.10) >DroEre_CAF1 14885 113 - 1 -----UUCGCCAAGUGCGCAUGUGUGCUUUUCUAUGAACGGUUAUGUAAGGCAUCCACUGUGCCGGCUUCUGCCCCAUUU-AAGCUG-AAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU -----...((((((.(((((....)))...........(((((......(((((.....)))))(((....)))......-.)))))-.......))))))))................. ( -27.70) >DroYak_CAF1 14405 113 - 1 -----UUCGCCAAGUGCGCAUGUGUGCUUUUCUAUGGUCGGUUAUGUAAGGCAUCCACUGUGCCGACUUCUGCCCCAUUU-UAGCUG-AAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU -----...((((((.(((.......(((.....((((..(((.......(((((.....))))).......)))))))..-.)))..-......)))))))))................. ( -27.70) >consensus A_AAUUUCGCCCAGUGCACAUGUGUGCUUUUCUAUGGUCGCUUAUGUAAGGCAACCACUGUGCCGGCUUCUGCCCCAUUUCAAGCUGGAAAACCCGCUUUGGCUUCGUUUUCCAUUUCAU ........((...(.(((((((..((((((..(((((....))))).))))))..)).))))))(((....))).........))((((((((..((....))...))))))))...... (-25.40 = -25.64 + 0.24)

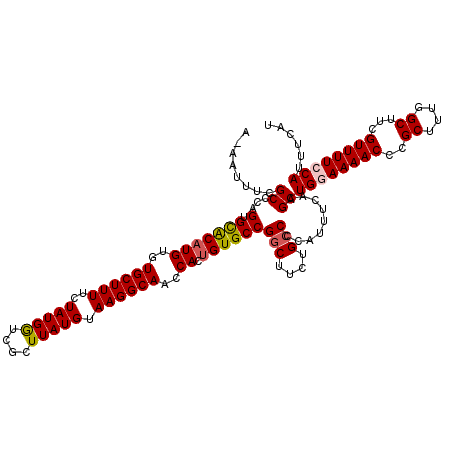
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:47 2006