| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,273,716 – 8,273,811 |
| Length | 95 |
| Max. P | 0.589508 |

| Location | 8,273,716 – 8,273,811 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -12.26 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
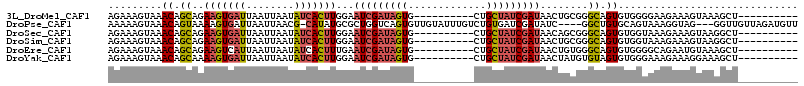
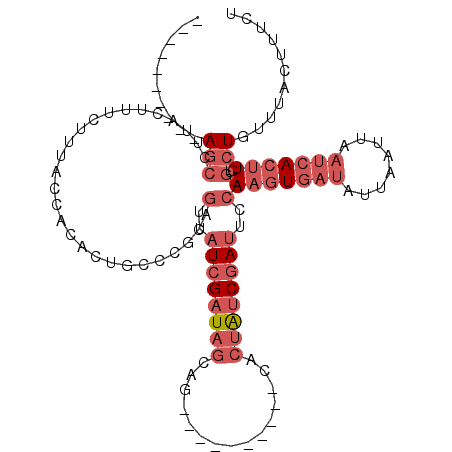
>3L_DroMel_CAF1 8273716 95 + 23771897 AGAAAGUAAACAGCAGAAGUGAUUAAUUAAUAUCACUUGGAAUCGAUAGUG----------CUGCUAUCGAUAACUGCGGGCAGUGUGGGGAAGAAAGUAAAGCU---------- .....((.....(((((((((((........)))))))...((((((((..----------...))))))))..))))..)).......................---------- ( -23.30) >DroPse_CAF1 7938 107 + 1 AAAAAGUAAACAGUAAAAGUGAUUAAUUAACG-CAUAUGCGCUGGUCAGUGUUGUAUUUGUCUGUGAUCGAUAUC----GGCUGUGCAGUAAAGGUAG---GGUUGUUAGAUGUU .........((((((((.(..((..((((.((-(....))).))))....))..).)))).)))).(((.....(----((((.(((.......))).---)))))...)))... ( -19.20) >DroSec_CAF1 9068 95 + 1 AGAAAGUAAACAGCAGAAGUGAUUAAUUAAUAUCACUUGGAAUCGAUAGUG----------CUGCUAUCGAUAACAGCGGGCAGUGUGGUAAAGAAAGUAAGGCU---------- ............((..(((((((........)))))))(..((((((((..----------...))))))))..).)).(((....((.(......).))..)))---------- ( -20.60) >DroSim_CAF1 9043 95 + 1 AGAAAGUAAACAGCAGAAGUGAUUAAUUAAUAUCACUUGGAAUCGAUAGUG----------CUGCUAUCGAUAACUGCGGGCAGUGUGGUAAAGAAAGUAAGGCU---------- ............(((((((((((........)))))))...((((((((..----------...))))))))..)))).(((....((.(......).))..)))---------- ( -23.40) >DroEre_CAF1 9173 95 + 1 AGAAAGUAAACAGCAGAAGUCAUUAAUUAAUAUCACUUUGAAUCGAUAGUG----------CUGCUAUCGAUAACUGUGGGCAGUGUGGGGCAGAAUGUAAAGCU---------- .....((..((((..(((((.((........)).)))))..((((((((..----------...))))))))..))))..))...((...((.....))...)).---------- ( -19.10) >DroYak_CAF1 9152 95 + 1 AGAAAGUAAACAGCAAAAGUGAUUAAUUAAUAUCACUUGGAAUCGAUAGUG----------CUGCUAUCGAUAACUAUGUGUAGUGUGGGAAAGAAAGGAAAGCU---------- ...........(((..(((((((........)))))))...((((((((..----------...)))))))).((((....)))).................)))---------- ( -19.70) >consensus AGAAAGUAAACAGCAGAAGUGAUUAAUUAAUAUCACUUGGAAUCGAUAGUG__________CUGCUAUCGAUAACUGCGGGCAGUGUGGGAAAGAAAGUAAAGCU__________ .........((.((..(((((((........)))))))...(((((((((.............)))))))))........)).)).............................. (-12.26 = -12.90 + 0.64)
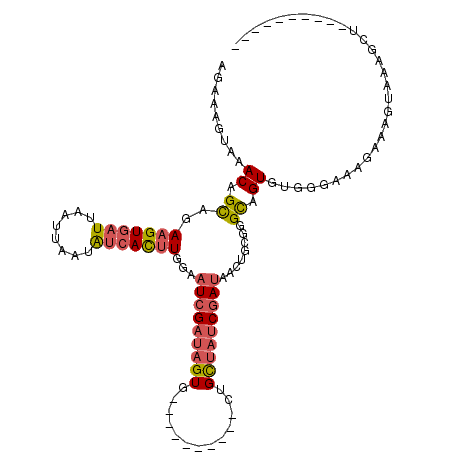

| Location | 8,273,716 – 8,273,811 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -15.97 |
| Consensus MFE | -7.25 |
| Energy contribution | -9.28 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8273716 95 - 23771897 ----------AGCUUUACUUUCUUCCCCACACUGCCCGCAGUUAUCGAUAGCAG----------CACUAUCGAUUCCAAGUGAUAUUAAUUAAUCACUUCUGCUGUUUACUUUCU ----------...........................((((..((((((((...----------..))))))))...(((((((........)))))))))))............ ( -18.90) >DroPse_CAF1 7938 107 - 1 AACAUCUAACAACC---CUACCUUUACUGCACAGCC----GAUAUCGAUCACAGACAAAUACAACACUGACCAGCGCAUAUG-CGUUAAUUAAUCACUUUUACUGUUUACUUUUU ..............---.............((((..----(((....)))......................(((((....)-))))...............))))......... ( -8.70) >DroSec_CAF1 9068 95 - 1 ----------AGCCUUACUUUCUUUACCACACUGCCCGCUGUUAUCGAUAGCAG----------CACUAUCGAUUCCAAGUGAUAUUAAUUAAUCACUUCUGCUGUUUACUUUCU ----------(((...........................(..((((((((...----------..))))))))..)(((((((........)))))))..)))........... ( -17.50) >DroSim_CAF1 9043 95 - 1 ----------AGCCUUACUUUCUUUACCACACUGCCCGCAGUUAUCGAUAGCAG----------CACUAUCGAUUCCAAGUGAUAUUAAUUAAUCACUUCUGCUGUUUACUUUCU ----------...........................((((..((((((((...----------..))))))))...(((((((........)))))))))))............ ( -18.90) >DroEre_CAF1 9173 95 - 1 ----------AGCUUUACAUUCUGCCCCACACUGCCCACAGUUAUCGAUAGCAG----------CACUAUCGAUUCAAAGUGAUAUUAAUUAAUGACUUCUGCUGUUUACUUUCU ----------(((..............(((((((....)))).((((((((...----------..)))))))).....))).((((....))))......)))........... ( -13.70) >DroYak_CAF1 9152 95 - 1 ----------AGCUUUCCUUUCUUUCCCACACUACACAUAGUUAUCGAUAGCAG----------CACUAUCGAUUCCAAGUGAUAUUAAUUAAUCACUUUUGCUGUUUACUUUCU ----------(((...........................(..((((((((...----------..))))))))..)(((((((........)))))))..)))........... ( -18.10) >consensus __________AGCCUUACUUUCUUUACCACACUGCCCGCAGUUAUCGAUAGCAG__________CACUAUCGAUUCCAAGUGAUAUUAAUUAAUCACUUCUGCUGUUUACUUUCU ..........(((...........................(..((((((((...............))))))))..)(((((((........)))))))..)))........... ( -7.25 = -9.28 + 2.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:26 2006