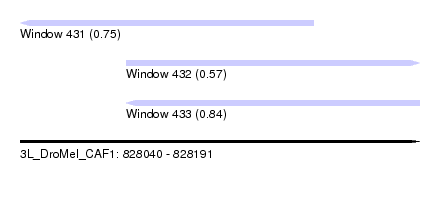
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 828,040 – 828,191 |
| Length | 151 |
| Max. P | 0.843851 |

| Location | 828,040 – 828,151 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -49.10 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
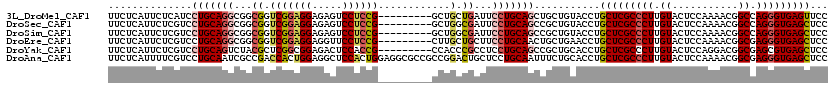
>3L_DroMel_CAF1 828040 111 - 23771897 UUCUCAUUCUCAUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCG---------GCUGCUGAUUCCUGCAGCUGCUGUACCUGCUCGCCCUUGUACUCCAAAACGGCCAGGGUGAGUUCC ..............((((((((((((((((((((.....))))))---------)))))))...)))))))...........(((((((((.((...........)).)))))))))... ( -51.80) >DroSec_CAF1 2324 111 - 1 UUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCG---------GCUGGCGAUUCCUGCAGCCGCUGUACCUGCUCGCCCUUGUACUCCAAAACGGCCAGGGUGAGCUCC ..........((..(((((((((.((((((((((.....))))))---------)))).))...))))))).))........(((((((((.((...........)).)))))))))... ( -52.00) >DroSim_CAF1 2373 111 - 1 UUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCG---------GCUGGCGAUUCCUGCAGCCGCUGUACCUGCUCGCCCUUGUACUCCAAAACGGCCAGGGUGAGCUCC ..........((..(((((((((.((((((((((.....))))))---------)))).))...))))))).))........(((((((((.((...........)).)))))))))... ( -52.00) >DroEre_CAF1 3345 111 - 1 UUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGGUUCCUCCG---------CUUGCUGCUUCCUGCAACUGCUGAACCUGCUCGCCCUUGUACUCCAAAACGGCGAGGGUGAGCUCC ......(((......((((((.((((((((((((.....))))))---------...)))))).))))))......)))...((((((((((((...........))))))))))))... ( -51.50) >DroYak_CAF1 2798 111 - 1 UUCUCAUUCUCGUCCUGCAGUCUACGCUCGGCGGAGACUCCACCG---------CCACCCGCCUCCUGCAGCCGCUGCACCUGCUCGCCCUUGUACUCCAGGACGGCGAGCGUGAGCUCC .........((((((((.((((...((..(((((........)))---------))....((....(((((...)))))...))..))....).))).)))))))).((((....)))). ( -40.20) >DroAna_CAF1 2963 120 - 1 UUCUCAUUUUCGUCCUGCAAUCGCCGACCACUGGAGGCUCCACUGGAGGCGCCGCCGGACUGCUCCUGCAAUUUCUGCACCUGCUCGCCCUUGUACUCCAAAACGGCGAGGGUGAGCUCC ...........((((.((...((((..(((.(((.....))).))).))))..)).))))......((((.....))))...((((((((((((...........))))))))))))... ( -47.10) >consensus UUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCG_________CCUGCCGAUUCCUGCAGCCGCUGUACCUGCUCGCCCUUGUACUCCAAAACGGCCAGGGUGAGCUCC ..............(((((((...((.(((((((.....))))))............).))...)))))))...........(((((((((.((...........)).)))))))))... (-30.12 = -30.82 + 0.70)
| Location | 828,080 – 828,191 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 828080 111 + 23771897 GUACAGCAGCUGCAGGAAUCAGCAGCCGGAGGACUCUCCUCCGACCGCCGCCUGCAGGAUGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAAGUUGGAGGACGAACGCGAU ...((((..(((((((.....((.(.((((((.....)))))).).))..))))))).((....)).....))))..(((..(.((((((...)))))).)....)))... ( -39.90) >DroSec_CAF1 2364 111 + 1 GUACAGCGGCUGCAGGAAUCGCCAGCCGGAGGACUCUCCUCCGACCGCCGCCUGCAGGACGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAACCUGGAGGACGAACGCGAU ...((((..(((((((....((..(.((((((.....)))))).).))..)))))))..(........)..))))..(((..(.((((((...)))))).)....)))... ( -41.50) >DroSim_CAF1 2413 111 + 1 GUACAGCGGCUGCAGGAAUCGCCAGCCGGAGGACUCUCCUCCGACCGCCGCCUGCAGGACGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAACCUGGAGGACGAACGCGAU ...((((..(((((((....((..(.((((((.....)))))).).))..)))))))..(........)..))))..(((..(.((((((...)))))).)....)))... ( -41.50) >DroEre_CAF1 3385 111 + 1 GUUCAGCAGUUGCAGGAAGCAGCAAGCGGAGGAACCUCCUCCGACCGCCGCCUGCAGGACGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAGGCUGGAGGACGAACGCGAU .((((((..(((((((..((.(....((((((.....)))))).).))..)))))))..(........)..))))))(((..(.((((((...)))))).)....)))... ( -42.40) >DroYak_CAF1 2838 111 + 1 GUGCAGCGGCUGCAGGAGGCGGGUGGCGGUGGAGUCUCCGCCGAGCGUAGACUGCAGGACGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAAGCUGGAGGACGAACGCGAU ...((((..((((((...(((..((((((........))))))..)))...))))))..(........)..))))..(((..(.((((((...)))))).)....)))... ( -42.60) >DroPer_CAF1 3306 99 + 1 GCGGCCCAACUACUAGAACAGGGUGGCAGU------------GGCGGACGGCUGCAGGGCGAGAACGAGAAGCUCAAGCGAAUGCUUCAGAGGCUGGAGGAUGAGCGCGAU ((.((((.............)))).)).(.------------.((.....))..).((((...........))))..(((.((.((((((...)))))).))...)))... ( -32.12) >consensus GUACAGCAGCUGCAGGAAUCGGCAGCCGGAGGACUCUCCUCCGACCGCCGCCUGCAGGACGAGAAUGAGAAGCUGAAGCGAAUGCUCCAGAAGCUGGAGGACGAACGCGAU ...((((..(((((((..........((((((.....)))))).......)))))))..(........)..))))..(((..(.((((((...)))))).)....)))... (-29.90 = -30.60 + 0.70)

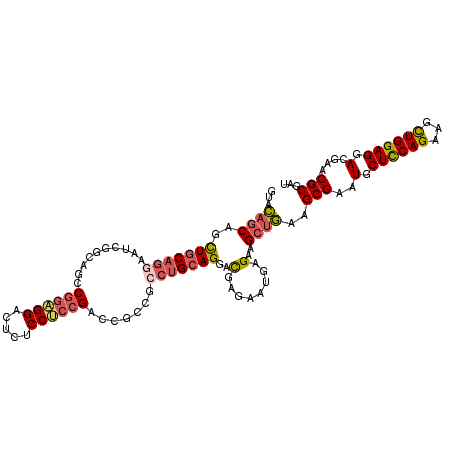
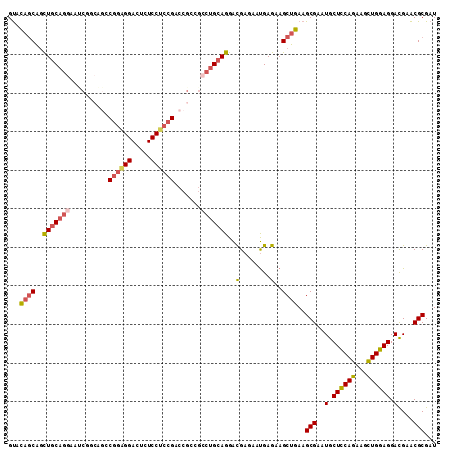
| Location | 828,080 – 828,191 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -41.41 |
| Consensus MFE | -27.04 |
| Energy contribution | -29.32 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 828080 111 - 23771897 AUCGCGUUCGUCCUCCAACUUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCAUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCGGCUGCUGAUUCCUGCAGCUGCUGUAC ...(((...((.(((((.....))))).)).)))..((((..............((((((((((((((((((((.....)))))))))))))...)))))))..))))... ( -48.99) >DroSec_CAF1 2364 111 - 1 AUCGCGUUCGUCCUCCAGGUUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCGGCUGGCGAUUCCUGCAGCCGCUGUAC ...(((..((......(((..(((((((....)))))))..)))......))..(((((((((.((((((((((.....)))))))))).))...))))))).)))..... ( -49.80) >DroSim_CAF1 2413 111 - 1 AUCGCGUUCGUCCUCCAGGUUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCGGCUGGCGAUUCCUGCAGCCGCUGUAC ...(((..((......(((..(((((((....)))))))..)))......))..(((((((((.((((((((((.....)))))))))).))...))))))).)))..... ( -49.80) >DroEre_CAF1 3385 111 - 1 AUCGCGUUCGUCCUCCAGCCUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGGUUCCUCCGCUUGCUGCUUCCUGCAACUGCUGAAC ...(((...((.((((((...)))))).)).)))((((((...............((((((.((((((((((((.....))))))...)))))).))))))...)))))). ( -45.77) >DroYak_CAF1 2838 111 - 1 AUCGCGUUCGUCCUCCAGCUUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCGUCCUGCAGUCUACGCUCGGCGGAGACUCCACCGCCACCCGCCUCCUGCAGCCGCUGCAC ...............((((..(((((((....)))))))...............((((((....((...(((((........)))))...))....))))))..))))... ( -32.60) >DroPer_CAF1 3306 99 - 1 AUCGCGCUCAUCCUCCAGCCUCUGAAGCAUUCGCUUGAGCUUCUCGUUCUCGCCCUGCAGCCGUCCGCC------------ACUGCCACCCUGUUCUAGUAGUUGGGCCGC .............((((((..(((.((((...((..((((.....))))..))...((((..(....).------------.)))).....)))).)))..)))))).... ( -21.50) >consensus AUCGCGUUCGUCCUCCAGCUUCUGGAGCAUUCGCUUCAGCUUCUCAUUCUCGUCCUGCAGGCGGCGGUCGGAGGAGAGUCCUCCGCCUGCCGAUUCCUGCAGCCGCUGUAC ...(((...((.((((((...)))))).)).)))..((((..............(((((((((.(((.((((((.....)))))).))).))...)))))))..))))... (-27.04 = -29.32 + 2.28)

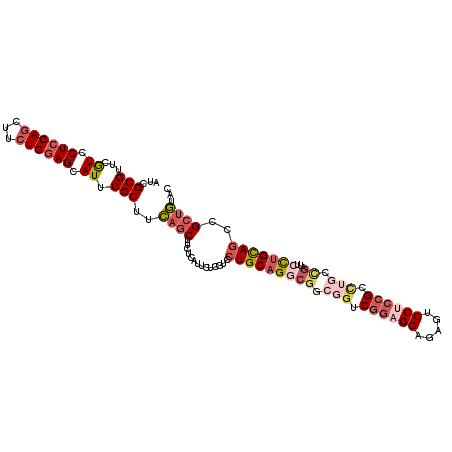
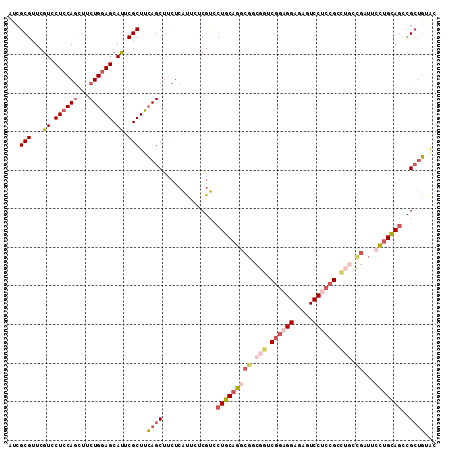
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:25 2006