
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,259,490 – 8,259,593 |
| Length | 103 |
| Max. P | 0.655764 |

| Location | 8,259,490 – 8,259,593 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -4.45 |
| Energy contribution | -5.07 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
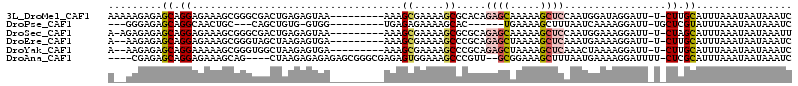
>3L_DroMel_CAF1 8259490 103 + 23771897 AAAAAGAGAGCAGGAGAAAGCGGGCGACUGAGAGUAA---------AAAGCGAAAAGCGCACAGAGCAAAAAGCUCCAAUGGAUAGGAUU-U-CUUGCAUUUAAAUAAUAAAUC .........((((((((...(((....))).......---------...(((.....)))...((((.....))))............))-)-)))))................ ( -21.30) >DroPse_CAF1 83127 91 + 1 ---GGGAGAGCAGGCAACUGC---CAGCUGUG-GUGG---------UGAGAGAAAAGCAC------UGAAAAGCUUUAAUCAAAAGGAUU-UGCUCGUAUUUAAAUAAUAAAUC ---....((((((((.((..(---((....))-)..)---------)..((..(((((..------......)))))..)).....).))-))))).................. ( -21.90) >DroSec_CAF1 81938 102 + 1 A-AGAGAGAGCAGGAGAAAGCGGGCGACUGAGAGUAA---------AAAGCGAAAAGCGCGCAGAGCAAAAAGCUCCAAUGGAAAGGAUU-U-CUAGCAUUUAAAUAAUAAAUU .-.......((.(((((..(((.((........((..---------...)).....)).))).((((.....))))............))-)-)).))................ ( -19.52) >DroEre_CAF1 86456 101 + 1 A--AAGAGAGCAGGAGAAAGCGGGUAGCUAAGAGUGA---------AAAGCGAAAAGCCCGCAGAGCUAAAAGCUCAAAUGAAAAGGAUU-U-CUUGCAUUUAAAUAAUAAAUC .--......((((((((..((((((.(((........---------..))).....)))))).((((.....))))............))-)-)))))................ ( -29.00) >DroYak_CAF1 86855 101 + 1 A--AAGAGAGCAGGAAAAAGCGGGUGGCUAAGAGUGA---------AAAGCGAAAAGCCCGCAGAGCUAAAAGCUCAAACUAAAAGGAUU-U-CUUGCAUUUAAAUAAUAAAUC .--......((((((((..((((((.(((........---------..))).....)))))).((((.....))))............))-)-)))))................ ( -28.90) >DroAna_CAF1 85131 103 + 1 ----CGAGAGCAGGAGAAAGCAG----CUAAGAGAGAGAGCGGGCGAGAGUGGAAAGCCCGUU--GCGGAAAGCUUUAAUGAAAAGGAUUUU-CUCGCAUUUAAAUAAUAAAUC ----.....((........))..----...........(((((((...........)))))))--((((((((((((......)))).))))-).)))................ ( -22.10) >consensus A__AAGAGAGCAGGAGAAAGCGGGCGGCUAAGAGUGA_________AAAGCGAAAAGCCCGCAGAGCAAAAAGCUCAAAUGAAAAGGAUU_U_CUUGCAUUUAAAUAAUAAAUC .........((.((...................................((.....)).....((((.....)))).................)).))................ ( -4.45 = -5.07 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:19 2006