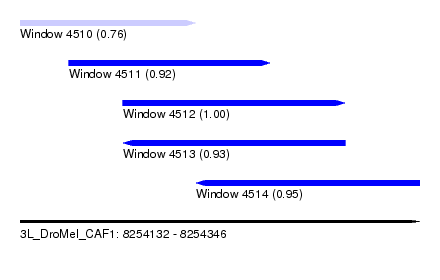
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,254,132 – 8,254,346 |
| Length | 214 |
| Max. P | 0.995594 |

| Location | 8,254,132 – 8,254,226 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -9.43 |
| Consensus MFE | -6.50 |
| Energy contribution | -6.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8254132 94 + 23771897 CACAUUCGU-ACAUCAUCUCGAUAAAA----------ACCAACAAACAACGUACA--AAUCCAUCGAA--UCCGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA .......((-((......(((((....----------....((.......))...--.....))))).--...))))...........(((((((.....)).))))). ( -11.19) >DroGri_CAF1 93492 94 + 1 CACAUUCAAAACUUCAC------AAAA-AAA---AAAAUCAACAACAAAAAAAAA--AUACCAUCGCA--U-CGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA .................------....-...---....................(--((((.......--.-.)))))..........(((((((.....)).))))). ( -7.00) >DroEre_CAF1 81140 93 + 1 CACAUUCGU-ACAUCAUCUCGAUAAAA----------ACCAACAAACAACGUACA--AAUCCAUCGAA--U-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA .......((-((......(((((....----------....((.......))...--.....))))).--.-.))))...........(((((((.....)).))))). ( -10.79) >DroWil_CAF1 98271 88 + 1 CACA------AAAAAAUCUC----AAA----------UCAAACAAACAACGUACAAAAAUCCAUCGAAAAU-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA ....------..........----...----------.............((((.....((....))....-.))))...........(((((((.....)).))))). ( -8.20) >DroYak_CAF1 81475 103 + 1 CACAUUCGU-ACAUCAUCACGAUAAAAAAAAAAACAAAUCAACAAACAACGUACA--AAUCCAUCGAA--U-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA .....((((-........))))............................((((.--..((....)).--.-.))))...........(((((((.....)).))))). ( -10.70) >DroAna_CAF1 79527 100 + 1 CUCAUUUCA-ACUUCAUCUCGAUAAAAAACCA---AAACCAACAAACAACGUACA--AAUCCAUCGAA--U-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA .........-......................---...............((((.--..((....)).--.-.))))...........(((((((.....)).))))). ( -8.70) >consensus CACAUUCGU_ACAUCAUCUCGAUAAAA__________ACCAACAAACAACGUACA__AAUCCAUCGAA__U_CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCA ........................................................................................(((((((.....)).))))). ( -6.50 = -6.50 + 0.00)

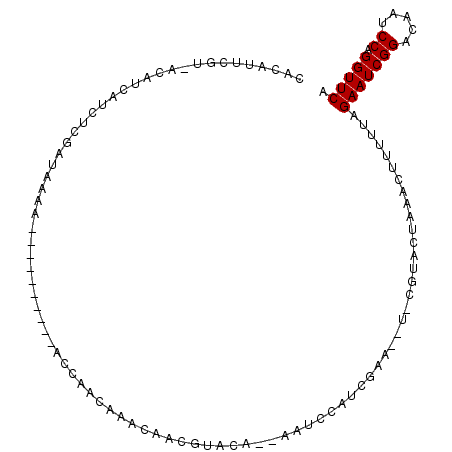
| Location | 8,254,158 – 8,254,266 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8254158 108 + 23771897 --------ACCAACAAACAACGUACA--AAUCCAUCGAA--UCCGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUCC --------.............((...--........(((--((.((..(((.....)))(((((((.....)).))))).)).))))).(((((((((.....))))..))))))).... ( -18.80) >DroPse_CAF1 77943 108 + 1 -----CAAACCAACAAACAACGUACA--ACUCCAUCGA---U-CGUACUAAACUU-UUAGAAUCGGACAAUCCAGGUUCCACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUAC -----................((((.--..........---.-.))))......(-(..(((((((.....)).(....)...)))))..))((((((.....))))))........... ( -17.52) >DroGri_CAF1 93514 112 + 1 AA---AAAAUCAACAACAAAAAAAAA--AUACCAUCGCA--U-CGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACUCUUCACAUAC ..---..((((..............(--((((.......--.-.)))))..........(((((((.....)).)))))....))))..(((((((((.....)))))))..))...... ( -17.60) >DroWil_CAF1 98288 111 + 1 --------UCAAACAAACAACGUACAAAAAUCCAUCGAAAAU-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACACUUCACAUAC --------.............((...........(((..(((-(((.....))......(((((((.....)).)))))....)))).))).((((((.....)))))).....)).... ( -17.40) >DroYak_CAF1 81503 115 + 1 AAAAACAAAUCAACAAACAACGUACA--AAUCCAUCGAA--U-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUCC .....................((...--........(((--(-(((.....))......(((((((.....)).)))))....))))).(((((((((.....))))..))))))).... ( -19.70) >DroMoj_CAF1 98322 95 + 1 --------------------AAAAAC--GAAACAUCGCA--U-UGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUAC --------------------......--........((.--.-............((..(((((((.....)).(......).)))))..))((((((.....))))))))......... ( -17.60) >consensus ________ACCAACAAACAACGUACA__AAUCCAUCGAA__U_CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUAC ............................................((.............(((((((.....)).)))))..........(((((((((.....))))..))))))).... (-15.18 = -15.23 + 0.06)

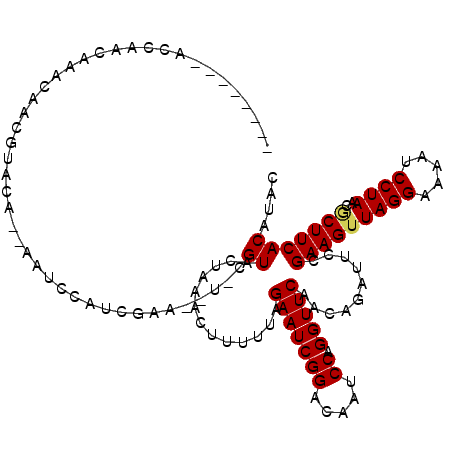
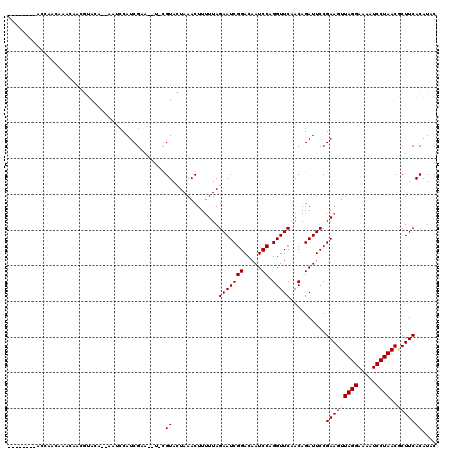
| Location | 8,254,187 – 8,254,306 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.30 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8254187 119 + 23771897 -UCCGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUCCCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA -......((((.....))))..((((((....((.((((....(....)(((((((((.....))))..)))))..............)))).))......(((....)))))))))... ( -24.30) >DroVir_CAF1 98577 118 + 1 -U-CGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA -(-((((((..........(((((((.....)).)))))..........(((((((((.....))))..)))))...............)))))))...(((((....)))))....... ( -26.60) >DroPse_CAF1 77974 117 + 1 -U-CGUACUAAACUU-UUAGAAUCGGACAAUCCAGGUUCCACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA -(-((((.......(-(..(((((((.....)).(....)...)))))..))((((((.....))))))......................)))))...(((((....)))))....... ( -24.10) >DroGri_CAF1 93548 118 + 1 -U-CGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACUCUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA -(-((((((..........(((((((.....)).)))))..........(((((((((.....)))))))..))...............)))))))...(((((....)))))....... ( -26.30) >DroWil_CAF1 98320 119 + 1 AU-CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACACUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA .(-((((............(((((((.....)).)))))..........(((((((((.....))))..))))).................)))))...(((((....)))))....... ( -24.30) >DroMoj_CAF1 98339 118 + 1 -U-UGUAUUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA -(-((((............(((((((.....)).)))))..........(((((((((.....))))..)))))..........)))))..........(((((....)))))....... ( -24.30) >consensus _U_CGUACUAAACUUUUUAGAAUCGGACAAUCCAGGUUCAACAGAUUCCGAAGUUAGGAAAAUCCUAACGCUUCACAUACCAAUUACAGAAUAUGACACACUCGGCUCCGAGUCCGACAA ......................((((((....((.((((....(....)(((((((((.....))))..)))))..............)))).))......(((....)))))))))... (-23.03 = -23.08 + 0.06)

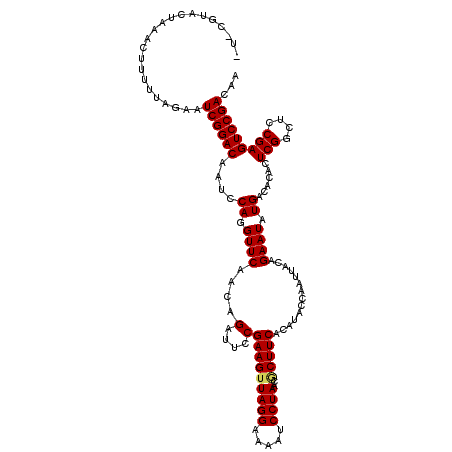
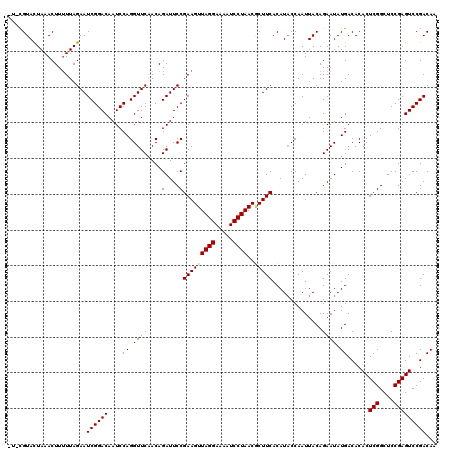
| Location | 8,254,187 – 8,254,306 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.30 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -30.34 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8254187 119 - 23771897 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGGAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAGUACGGA- ..(((((((......(((((.((.(((((((((......))))))))).))((((((.....)))))))))))(((((.........))))))))))))....................- ( -36.00) >DroVir_CAF1 98577 118 - 1 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAAUACG-A- ..(((((((......(((((.((.((((((.((......)).)))))).))((((((.....)))))))))))(((((.........))))))))))))..................-.- ( -32.10) >DroPse_CAF1 77974 117 - 1 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGUGGAACCUGGAUUGUCCGAUUCUAA-AAGUUUAGUACG-A- ..(((((((......(((((.((.((((((.((......)).)))))).))((((((.....)))))))))))((((((.(....)))))))))))))).....-............-.- ( -33.10) >DroGri_CAF1 93548 118 - 1 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGAGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAAUACG-A- ..(((((((......((((.....((((((.((......)).))))))..(((((((.....)))))))))))(((((.........))))))))))))..................-.- ( -30.40) >DroWil_CAF1 98320 119 - 1 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGUGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAGUACG-AU ..(((((((......(((((....((((((.((......)).))))))...((((((.....)))))))))))(((((.........))))))))))))..................-.. ( -29.80) >DroMoj_CAF1 98339 118 - 1 UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAAUACA-A- ..(((((((......(((((.((.((((((.((......)).)))))).))((((((.....)))))))))))(((((.........))))))))))))..................-.- ( -32.10) >consensus UUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGUUGAACCUGGAUUGUCCGAUUCUAAAAAGUUUAAUACG_A_ ..(((((((......(((((.((.((((((.((......)).)))))).))((((((.....)))))))))))(((((.........))))))))))))..................... (-30.34 = -30.40 + 0.06)

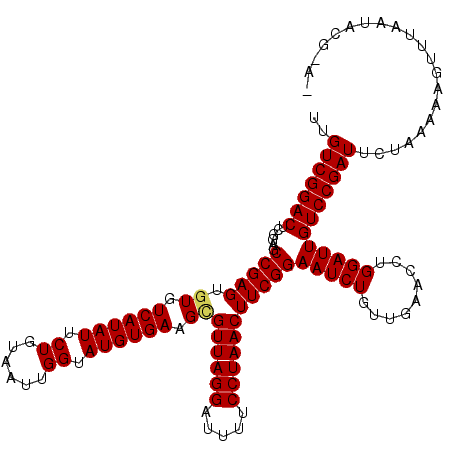
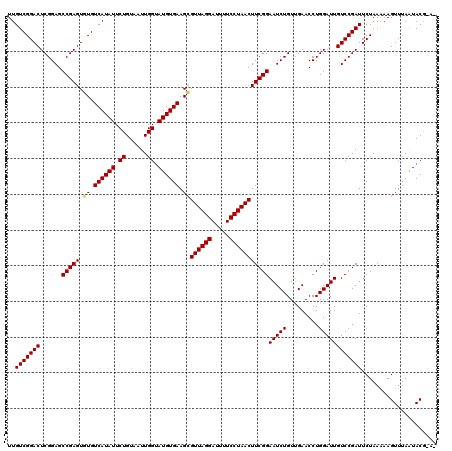
| Location | 8,254,226 – 8,254,346 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -35.84 |
| Energy contribution | -35.62 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8254226 120 - 23771897 UCGAUACUUGGCCGAGUUUAGAGCUCGCGCAGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGGAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGU ((((.((((((((((((((...((....)).((((......)))))))))))..)))))))((.(((((((((......))))))))).))((((((.....)))))).))))....... ( -43.00) >DroVir_CAF1 98615 120 - 1 GCGAUAUUUGGCCGAGUUAAGAGCGCGCGCGGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGU ((.(((((((((((((((....((....)).((((......)))).))))))..))))))))).((((((.((......)).)))))).))((((((.....))))))............ ( -37.90) >DroGri_CAF1 93586 120 - 1 GCGAUAUUUGGCCGAGUUAAGAGCGCGUGCAGACACAUCGUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGAGUUAGGAUUUUCCUAACUUCGGAAUCUGU .(((((((((((((((((.(.((((.(((....)))..)))).)..))))))..))))))))).((((((.((......)).)))))).((((((((.....))))))))))........ ( -36.90) >DroWil_CAF1 98359 120 - 1 GCGAUAUUUGGCCGAGUUAAGAGCGCGUGCAGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGUGUUAGGAUUUUCCUAACUUCGGAAUCUGU .(((((((((((((((((....((....)).((((......)))).))))))..))))))))).((((((.((......)).))))))...((((((.....))))))..))........ ( -34.60) >DroMoj_CAF1 98377 120 - 1 GCGAUAUUUGGCCGAGUUAAGAGCGCGGGCGGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGU ((.(((((((((((((((....((....)).((((......)))).))))))..))))))))).((((((.((......)).)))))).))((((((.....))))))............ ( -37.50) >DroAna_CAF1 79627 120 - 1 GCGAUACUUGGCCGAGUUUAGCGCGCGUGCAGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGGAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGU ((.((((((((((((((((.(((....))).((((......)))))))))))..))))))))).(((((((((......))))))))).))((((((.....))))))............ ( -45.60) >consensus GCGAUAUUUGGCCGAGUUAAGAGCGCGUGCAGACACAUCAUUGUCGGACUCGGAGCCGAGUGUGUCAUAUUCUGUAAUUGGUAUGUGAAGCGUUAGGAUUUUCCUAACUUCGGAAUCUGU .(((((((((((((((((....((....)).((((......)))).))))))..))))))))).((((((.((......)).))))))...((((((.....))))))..))........ (-35.84 = -35.62 + -0.22)

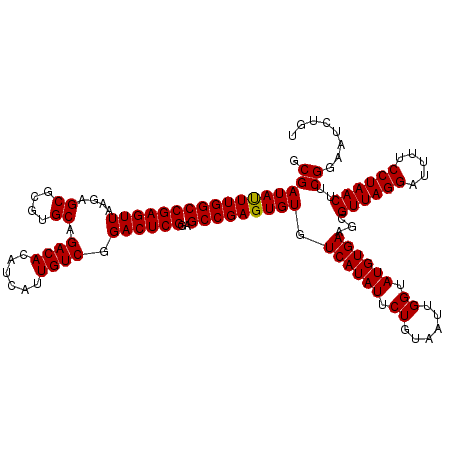
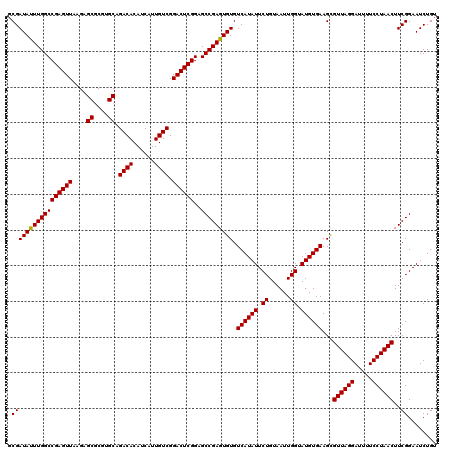
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:17 2006