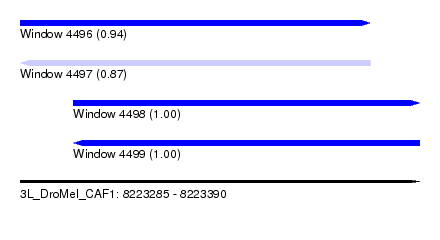
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,223,285 – 8,223,390 |
| Length | 105 |
| Max. P | 0.999773 |

| Location | 8,223,285 – 8,223,377 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -19.48 |
| Energy contribution | -20.52 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8223285 92 + 23771897 GAUUCUCGAAAGGGGUUGUGUGUGGUUGGUUGGUGCUUUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA ....(((....)))(((((((.(((((((..((((((((....((....))...))))))))))))))))))))))(((.....)))..... ( -26.70) >DroSec_CAF1 45781 90 + 1 GAUUCUCGAAAGGGGUUGU--GUGGUUGGUCGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA ...((((....)))).(((--(((.(((...((((.((((.((((((((.((.....)).))))))))..)))).))))...))).)))))) ( -29.50) >DroSim_CAF1 43696 90 + 1 GAUUCUCGAAAGGGGUUGU--GUGGUUGGUUGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA ...((((....)))).(((--(((.(((..(((((.((((.((((((((.((.....)).))))))))..)))).)))))..))).)))))) ( -31.00) >DroEre_CAF1 49103 80 + 1 GA--CUCGA--GGGGUUUG--GCG------UGGGGCCGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA ((--((...--..))))((--(((------(...((.((..((((((((.((.....)).)))))))).)))).))))))............ ( -24.20) >DroYak_CAF1 47390 83 + 1 GAUUCUCGA-UGGGGUUGU--GUG------UGGGCCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCACAUAGGUGCAUA ........(-((.(.((((--(((------.(.(..((((.((((((((.((.....)).))))))))..)))).).)))))))).).))). ( -24.90) >consensus GAUUCUCGAAAGGGGUUGU__GUGGUUGGUUGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA ....(((....))).......(((......(((((.((((.((((((((.((.....)).))))))))..)))).)))))......)))... (-19.48 = -20.52 + 1.04)


| Location | 8,223,285 – 8,223,377 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8223285 92 - 23771897 UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCAAAGCACCAACCAACCACACACAACCCCUUUCGAGAAUC ............(((.(((.....((((((((.((.....)).))))))))....))).))).............................. ( -13.60) >DroSec_CAF1 45781 90 - 1 UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCGACCAACCAC--ACAACCCCUUUCGAGAAUC ............(((((.(((((.((((((((.((.....)).))))))))..)).))).)).))).....--................... ( -16.80) >DroSim_CAF1 43696 90 - 1 UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCAACCAACCAC--ACAACCCCUUUCGAGAAUC ............(((.((((.(..((((((((.((.....)).)))))))).).)))).))).........--................... ( -16.30) >DroEre_CAF1 49103 80 - 1 UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACGGCCCCA------CGC--CAAACCCC--UCGAG--UC ............((((((.((((.((((((((.((.....)).))))))))..)).))...)------)))--))......--.....--.. ( -18.40) >DroYak_CAF1 47390 83 - 1 UAUGCACCUAUGUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGGCCCA------CAC--ACAACCCCA-UCGAGAAUC ..........(((((.(((.....((((((((.((.....)).)))))))).....))))))------)).--.........-......... ( -17.00) >consensus UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCAACCAACCAC__ACAACCCCUUUCGAGAAUC ............(((.((((.(..((((((((.((.....)).)))))))).).)))).))).............................. (-13.88 = -13.96 + 0.08)


| Location | 8,223,299 – 8,223,390 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.80 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8223299 91 + 23771897 GUUGUGUGUGGUUGGUUGGUGCUUUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUA----UAUGCACAUAUAU (((((((.(((((((..((((((((....((....))...))))))))))))))))))))))...(((((.(((((..----..))))).))))) ( -27.90) >DroSec_CAF1 45795 93 + 1 GUUGU--GUGGUUGGUCGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUAUACAUAUGCACAUAUAU .....--..........((((.((((.((((((((.((.....)).))))))))..)))).))))(((((.(((((((....))))))).))))) ( -30.10) >DroSim_CAF1 43710 93 + 1 GUUGU--GUGGUUGGUUGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUAUACAUAUGCACAUAUUU .((((--(((((.(.....(((.((..((((((((.((.....)).)))))))).))))).))))))))))(((((((....)))))))...... ( -30.00) >DroEre_CAF1 49113 83 + 1 GUUUG--GCG------UGGGGCCGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUAUA--U--GCACAUAUAU ...((--(((------(...((.((..((((((((.((.....)).)))))))).)))).))))))((((.(((((....--)--)))).)))). ( -30.40) >DroYak_CAF1 47403 85 + 1 GUUGU--GUG------UGGGCCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCACAUAGGUGCAUAUA--UGUGCACAUGUAU ....(--(((------(((...((((.((((((((.((.....)).))))))))..))))...))))))).((((((...--.))))))...... ( -29.30) >consensus GUUGU__GUGGUUGGUUGGUGCUGUGGAAUUGAAAAGCGAAAAGCAUUUCAAUUAACGCAACGCCAUAUAGGUGCAUAUA__UAUGCACAUAUAU .................((((.((((.((((((((.((.....)).))))))))..)))).))))(((((.(((((((....))))))).))))) (-23.28 = -24.80 + 1.52)


| Location | 8,223,299 – 8,223,390 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8223299 91 - 23771897 AUAUAUGUGCAUA----UAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCAAAGCACCAACCAACCACACACAAC .((((.(((((..----..))))).))))(((.(((.....((((((((.((.....)).))))))))....))).)))................ ( -21.10) >DroSec_CAF1 45795 93 - 1 AUAUAUGUGCAUAUGUAUAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCGACCAACCAC--ACAAC .((((.(((((((....))))))).))))(((((.(((((.((((((((.((.....)).))))))))..)).))).)).))).....--..... ( -26.70) >DroSim_CAF1 43710 93 - 1 AAAUAUGUGCAUAUGUAUAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCAACCAACCAC--ACAAC ..(((.(((((((....))))))).))).(((.((((.(..((((((((.((.....)).)))))))).).)))).))).........--..... ( -25.00) >DroEre_CAF1 49113 83 - 1 AUAUAUGUGC--A--UAUAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACGGCCCCA------CGC--CAAAC .((((.((((--(--....))))).))))((((((.((((.((((((((.((.....)).))))))))..)).))...)------)))--))... ( -25.90) >DroYak_CAF1 47403 85 - 1 AUACAUGUGCACA--UAUAUGCACCUAUGUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGGCCCA------CAC--ACAAC ......(((((..--....)))))...(((((.(((.....((((((((.((.....)).)))))))).....))))))------)).--..... ( -22.30) >consensus AUAUAUGUGCAUA__UAUAUGCACCUAUAUGGCGUUGCGUUAAUUGAAAUGCUUUUCGCUUUUCAAUUCCACAGCACCAACCAACCAC__ACAAC (((((.(((((((....))))))).)))))((.((((.(..((((((((.((.....)).)))))))).).)))).))................. (-20.60 = -21.72 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:03 2006