
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,214,917 – 8,215,038 |
| Length | 121 |
| Max. P | 0.881139 |

| Location | 8,214,917 – 8,215,008 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 73.18 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8214917 91 - 23771897 GUGCACCACCAGCUAC--------AUUC---CACCCAACACC-CACUAGCCACCCC--UCCCACCUGGCUGUGGAUGCUGUGCGCAAGUGCGCAGAAUCCGCA--UC ((((............--------....---.......((.(-(((.(((((....--.......))))))))).))(((((((....))))))).....)))--). ( -26.30) >DroSec_CAF1 37100 91 - 1 GUGCACCACCAGCUCC--------AUUC---CAUCCACCACCCCACCAGUCCUCUC--AACCACCUGGCUGUGGAUGCUGUG-GCAAUUGCGCAGAAUCCGCA--UC (((((((((.((((((--------((.(---((.......................--.......)))..))))).))))))-)....)))))..........--.. ( -21.81) >DroSim_CAF1 35104 92 - 1 GUGCACCACCAGCUCC--------AUUC---CAUCCACCACCCCACUAGCCCCCUC--CUCCACCUGGCUGUGGAUGCUGUGCGCAAGUGCGCAGAAUCCGCA--UC ((((............--------....---(((((((.........((((.....--........)))))))))))(((((((....))))))).....)))--). ( -25.42) >DroEre_CAF1 40716 102 - 1 GUGCACCACCAGCGCCCCCUCGACCUUCCACCAUCCACCACC-CACUGGACUCGCCA-CCCCACCUGGCUGUGGAUGCUGGA-GCUGAUGCGCAGAAUCCGCA--UC ((((.......))))..........(((((.(((((((..((-....))....((((-.......)))).))))))).))))-)..((((((.......))))--)) ( -31.40) >DroYak_CAF1 38937 79 - 1 GUGCACCACC--------------------------CCCAAC-CAAUGGAUUCGCCAUCCCCACCUGGCUGUGGAUGCUGGA-AGUGAUGCGCAGAAUCCGCAAAUC (((((.(((.--------------------------.(((..-...))).((((.(((((.((......)).))))).))))-.))).))))).............. ( -21.20) >consensus GUGCACCACCAGCUCC________AUUC___CAUCCACCACC_CACUAGACCCCCC__CCCCACCUGGCUGUGGAUGCUGUG_GCAAAUGCGCAGAAUCCGCA__UC ...............................................((((...............))))((((((.(((((........))))).))))))..... (-12.78 = -13.50 + 0.72)



| Location | 8,214,946 – 8,215,038 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8214946 92 - 23771897 UGCAGGUUGCCAUAGCGGCCACAUCGCCAUGUGCACCACCAGCUAC--------AUUC---CACCCAACACC-CACUAGCCACCCC-UCCCACCUGGCUGUGGAU .((.(((((((((.((((.....)))).))).)))..))).))...--------....---..........(-(((.(((((....-.......))))))))).. ( -25.90) >DroSec_CAF1 37128 93 - 1 UGCAGGUUACCAUAGUGGCCACAUCGCCAUGUGCACCACCAGCUCC--------AUUC---CAUCCACCACCCCACCAGUCCUCUC-AACCACCUGGCUGUGGAU .((.(((...((((.((((......))))))))....))).))...--------....---......((((....((((.......-......))))..)))).. ( -19.62) >DroSim_CAF1 35133 93 - 1 UGCAGGUUGCCAUAGUGGCCACAUCGCCAUGUGCACCACCAGCUCC--------AUUC---CAUCCACCACCCCACUAGCCCCCUC-CUCCACCUGGCUGUGGAU .((.((((((....(((((......)))))..)))..))).))...--------....---...........((((.((((.....-........)))))))).. ( -23.92) >DroEre_CAF1 40744 104 - 1 UGCAGGUUGCCAUGGUGGCCACAUCGCCAUGUGCACCACCAGCGCCCCCUCGACCUUCCACCAUCCACCACC-CACUGGACUCGCCACCCCACCUGGCUGUGGAU .((.((((((((((((((.....)))))))).)))..))).)).............(((((..((((.....-...))))...((((.......)))).))))). ( -33.00) >consensus UGCAGGUUGCCAUAGUGGCCACAUCGCCAUGUGCACCACCAGCUCC________AUUC___CAUCCACCACC_CACUAGCCCCCCC_CCCCACCUGGCUGUGGAU (((.((((((....))))))((((....)))))))((((.(((.....................................................))))))).. (-17.00 = -16.62 + -0.38)

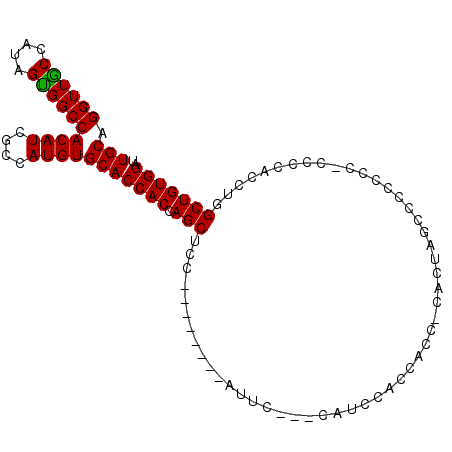
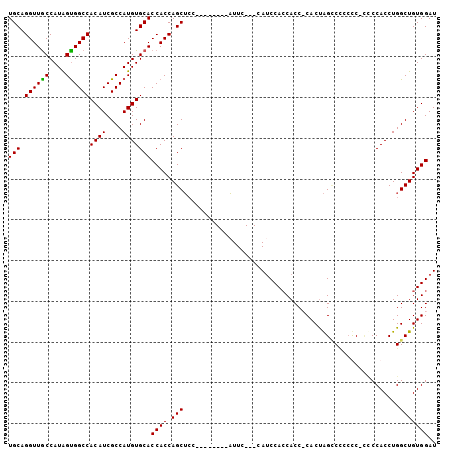
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:54 2006