
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,202,565 – 8,202,680 |
| Length | 115 |
| Max. P | 0.710333 |

| Location | 8,202,565 – 8,202,680 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -28.11 |
| Energy contribution | -28.08 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
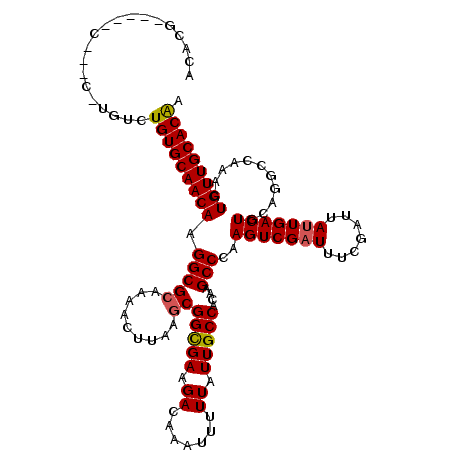
>3L_DroMel_CAF1 8202565 115 + 23771897 ACCCA-----GGACCUUGUGUGUGCAACAAGGCGCACAACUUAAGCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGAUUUCGAUUAUUGACUGCAGGCCAAACUGUUGCACAA .....-----((..(((((((((((......)))))).........(((((.((.......)).)))))))))).)).(((((((.......)))))))((((......))))....... ( -30.20) >DroPse_CAF1 29737 111 + 1 AUACG-----A---GUGCUGUGUGCAACAAGGCGCAGAACUUAAGCGGUGAAGACAAAUUUUUAUUGCCGCAAGCCCGAGUCGAUUUCGAUUAUUGACUG-GGGCCAAACUGUUGCACGA .....-----.---......(((((((((..(((..........((((((((((....)))))))))))))..((((.(((((((.......))))))).-)))).....))))))))). ( -39.00) >DroGri_CAF1 34596 113 + 1 GCACG-----C-UGCCUGUCUGUGCAACAAGGCGCAAAACUUAAGCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGUUUUCGAUUAUUGACUGCAAG-CAAACUGUUGCACAC ((...-----.-.)).....(((((((((.(((((.........))(((((.((.......)).)))))....)))..(((((....)))))............-.....))))))))). ( -27.40) >DroWil_CAF1 38579 113 + 1 ACUGU-----CGAC--UGUCUGUGCAACAAGGCGCAAAACUUAAGCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGAUUUCGAUUAUUGACUGCAGGCCAAACUGUUGCACAC .....-----....--....(((((((((..((...........))(((((.((.......)).)))))....(((..(((((((.......)))))))...))).....))))))))). ( -29.80) >DroMoj_CAF1 28437 118 + 1 GGGCGUCUGUC-UGUCUGUCUGUGCAACAAGGCGCAAAACUUAAGCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGAUUUCGAUUAUUGACUGCAAG-CAAACUGUUGCACAG (((((......-)))))..((((((((((.(((((.........))(((((.((.......)).)))))....)))..(((((((.......))))))).....-.....)))))))))) ( -34.40) >DroAna_CAF1 28485 102 + 1 --------------C----GUGUGCAACAAGGCGCACAACUUAACCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGAUUUCGAUUAUUGACUGCAGGCCAAACUGUUGCACGA --------------.----.(((((((((.................(((((.((.......)).)))))....(((..(((((((.......)))))))...))).....))))))))). ( -29.20) >consensus ACACG_____C___C_UGUCUGUGCAACAAGGCGCAAAACUUAAGCGGCGAAGACAAAUUUUUAUUGCCACAAGCCCAAGUCGAUUUCGAUUAUUGACUGCAGGCCAAACUGUUGCACAA ....................(((((((((.(((((.........))(((((.((.......)).)))))....)))..(((((((.......)))))))...........))))))))). (-28.11 = -28.08 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:51 2006