
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,195,306 – 8,195,402 |
| Length | 96 |
| Max. P | 0.759131 |

| Location | 8,195,306 – 8,195,402 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
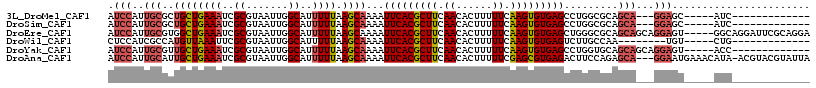
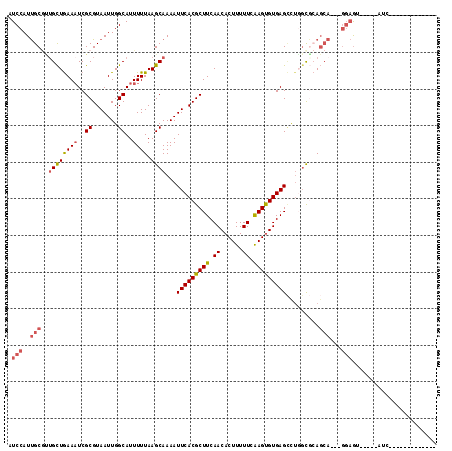
>3L_DroMel_CAF1 8195306 96 + 23771897 AUCCAUUGCGCUGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGCCUGGCGCAGCA---GGAGC-----AUC------------- ......(((.((((((....)((((....((((((((.....)))))((((((((.((......)).)))))))))))..))))))))---)..))-----)..------------- ( -27.50) >DroSim_CAF1 20870 96 + 1 AUCCAUUGCGCUGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGCCUGGCGCAGCA---GGAGC-----AUC------------- ......(((.((((((....)((((....((((((((.....)))))((((((((.((......)).)))))))))))..))))))))---)..))-----)..------------- ( -27.50) >DroEre_CAF1 26298 112 + 1 AUCCAUUGCGUGGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGCUGGGCGCAGCAGCAGGAGU-----GGCAGGAUUCGCAGGA (((((((((((((......)))))))))((.(((((((..((.....((((((((.((......)).))))))))((((....)))).))))))))-----).))))))........ ( -33.60) >DroWil_CAF1 24283 91 + 1 CUCCAUCGCCAUGUUAAAUUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGUCUUGCCAA--------UGU-----CUG------------- .......((((.((((........))))))))........((((.((((((((((.((......)).)))))))))).))))...--------...-----...------------- ( -21.20) >DroYak_CAF1 23750 99 + 1 AUCCAUUGCGUUGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGCCUGGUGCAGCAGCAGGAGU-----ACC------------- .(((.....(((((((..(((((((((((.((........))..)))).))))((((.(((((....)))))))))...))).))))))).)))..-----...------------- ( -24.70) >DroAna_CAF1 22012 113 + 1 AUCCAUUGCAUUGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCGAGCGUGAGACUUCCAGAGCA---GGAAUGAAACAUA-ACGUACGUAUUA .(((..(((.(((((((((..((.......))..)))).)))))..(((((((((.((......)).))))))))).........)))---)))..........-............ ( -24.10) >consensus AUCCAUUGCGUUGCUGAAAUCGCGUAAUUGGCAUUUUUAAGCAAAAUUCACGCUUCAACACUUUUUCAAGUGUGAGCCUGGCGCAGCA___GGAGU_____AUC_____________ .(((..(((..((((((((..((.......))..)))).))))...(((((((((.((......)).))))))))).........)))...)))....................... (-16.74 = -17.52 + 0.78)

| Location | 8,195,306 – 8,195,402 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
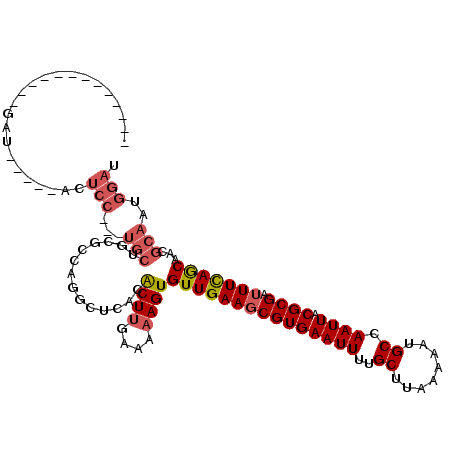
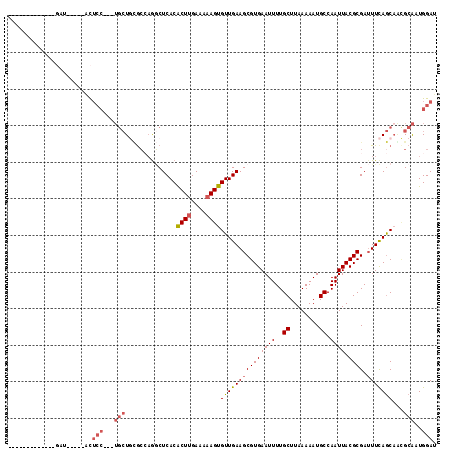
>3L_DroMel_CAF1 8195306 96 - 23771897 -------------GAU-----GCUCC---UGCUGCGCCAGGCUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCAGCGCAAUGGAU -------------...-----..(((---...(((((...((..((((((....))))))(((((((((((((..((........)).)))).)))).))))))).)))))..))). ( -30.00) >DroSim_CAF1 20870 96 - 1 -------------GAU-----GCUCC---UGCUGCGCCAGGCUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCAGCGCAAUGGAU -------------...-----..(((---...(((((...((..((((((....))))))(((((((((((((..((........)).)))).)))).))))))).)))))..))). ( -30.00) >DroEre_CAF1 26298 112 - 1 UCCUGCGAAUCCUGCC-----ACUCCUGCUGCUGCGCCCAGCUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCCACGCAAUGGAU (((((((......((.-----......)).(((((((...((((((((((....))))).)).)))((((..((.((........)).)))))))))....))))..))))..))). ( -27.40) >DroWil_CAF1 24283 91 - 1 -------------CAG-----ACA--------UUGGCAAGACUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAAUUUAACAUGGCGAUGGAG -------------...-----.((--------(((.((....((((((((....))))).))).((((((..((.((........)).))))))))..........)).)))))... ( -23.80) >DroYak_CAF1 23750 99 - 1 -------------GGU-----ACUCCUGCUGCUGCACCAGGCUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCAACGCAAUGGAU -------------...-----..(((((((((((........((((((((....))))).))).((((((..((.((........)).)))))))).....)))))..)))..))). ( -26.30) >DroAna_CAF1 22012 113 - 1 UAAUACGUACGU-UAUGUUUCAUUCC---UGCUCUGGAAGUCUCACGCUCGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCAAUGCAAUGGAU ............-.....(((((((.---((((...(((((((((((((......)))).))).((((((..((.((........)).))))))))))))))))))..).)))))). ( -23.40) >consensus _____________GAU_____ACUCC___UGCUGCGCCAGGCUCACACUUGAAAAAGUGUUGAAGCGUGAAUUUUGCUUAAAAAUGCCAAUUACGCGAUUUCAGCAACGCAAUGGAU .......................(((...(((..............((((....))))(((((((((((((((..((........)).)))).)))).)))))))...)))..))). (-17.92 = -18.83 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:42 2006