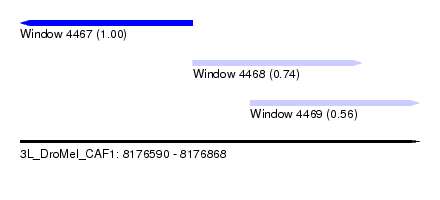
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,176,590 – 8,176,868 |
| Length | 278 |
| Max. P | 0.998184 |

| Location | 8,176,590 – 8,176,710 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -41.14 |
| Consensus MFE | -37.04 |
| Energy contribution | -38.16 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
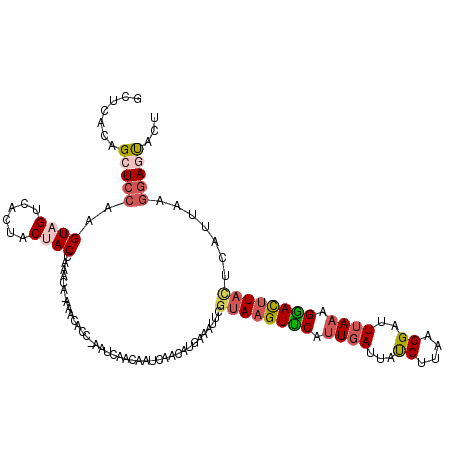
>3L_DroMel_CAF1 8176590 120 - 23771897 ACAACCCACUUGGUCAUCCGGCGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGUCGACAUGGCAGCACAACGUCUUUGACUCCGAUCCAACUUACUGGCCAGUUGAAGACGUUGUUGCU ............(((((.((((((.(((.((((........)).)).))).))))))..)))))(((((((((((((..(((.....(((......)))..)))..))))))))).)))) ( -45.70) >DroSec_CAF1 4371 120 - 1 ACAAACAACUUGGUCAUCCGGCGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGCCGACAUGGCAGCACAACGUCUUUGACUCCGAUCCAACUUACUGGCCAGUUGAAGAAGUUGUUGCU ............(((((.((((((.(((.((((........)).)).))).))))))..)))))((((((((.((((..(((.....(((......)))..)))..)))).)))).)))) ( -42.70) >DroSim_CAF1 4346 120 - 1 ACAAACCACUUGGUCAUCCGGCGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGCCGACAUGGCAGCACAACGUCUUUGACUCCGAUCCAACUUACUGGCCAGUUGAAGACGUUGUUGCU ............(((((.((((((.(((.((((........)).)).))).))))))..)))))(((((((((((((..(((.....(((......)))..)))..))))))))).)))) ( -48.40) >DroEre_CAF1 4574 120 - 1 ACACCUCACUUGGCCAUCUGGAGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGUCGACAUCGCAGCACAAAGUCUUUGACUCCGAUCCGAGUUACUUGCCAAUUGAAGACGUUGUUGCU ...........((((.((....)).))))((((........)).))(((((....)))))..((((((....(((((((((((......)))))...........))))))..)))))). ( -33.60) >DroYak_CAF1 4455 120 - 1 ACACCUCACUUGGUCAUCCGGCGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGCCGACGUCGCAGCACAAAGUCUUUGACUCCGAUCCAACUUGCUUGCCAAUUGAAGACGUUGUUGCU ..(((......)))....((((((.(((.((((........)).)).))).)))))).....((((((....(((((..(...(((.(......).)))....)..)))))..)))))). ( -35.30) >consensus ACAACCCACUUGGUCAUCCGGCGAAGGCCGCGCUUACACAUGCAGCUGUCGUCGCCGACAUGGCAGCACAACGUCUUUGACUCCGAUCCAACUUACUGGCCAGUUGAAGACGUUGUUGCU ............(((((.((((((.(((.((((........)).)).))).))))))..)))))(((((((((((((..(((.....(((......)))..)))..))))))))).)))) (-37.04 = -38.16 + 1.12)


| Location | 8,176,710 – 8,176,828 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -14.98 |
| Energy contribution | -16.18 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8176710 118 + 23771897 AUAUAAGCUGGCCAGCUGGGUUAAUUCAACAUCAAACCUAGCUCACAGCUCCCAGUAGUCACUUCUACCAAACA-AAGCACC-AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAU .....(((((...(((((((((............)))))))))..)))))....((((......))))......-.......-......((((.((((((.....)))..))).)))).. ( -24.40) >DroSec_CAF1 4491 112 + 1 AUAUAAGCUGGCUAGCUGGGUUAACUCGACAUCAAACCAAGCUCACAGCUCCAAGUAGUCACUACAACCAAACACAAAAACC-AAU-------CAACAUGAAAUUCGUAAGUUCAUUGAU .....(((((...((((.((((............)))).))))..)))))....((((...)))).................-.((-------((..(((((...(....)))))))))) ( -19.20) >DroSim_CAF1 4466 119 + 1 AUAUAAGCUGGCCAGCUGGGUUAACUCGACAUCAAACCAAGCUUACAGCUCCAAGUAGUCACUACAACCAAACACAAAAACC-AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAU .....(((((...((((.((((............)))).))))..)))))....((((...)))).................-......((((.((((((.....)))..))).)))).. ( -19.30) >DroEre_CAF1 4694 115 + 1 AUAUAAACUGGCCAGCUUGGUUGAGUCAGUAUCAAACCAAGCUCACAGCUCCAAGUAGUCACUGCUACCAAACA-AAACCCC-AUUCAACAAUC---AUGAAAUUCGUAAGUCCAUUGAU .......(((...(((((((((((.......)).)))))))))..)))......(((((....)))))......-.......-........(((---(.......(....).....)))) ( -20.60) >DroYak_CAF1 4575 116 + 1 AUAUAAACUGGCCAGCUGGGUUAACUCAAUAUCAAACACAGCUCACAGCUUCAAGUAGUCACUACUACCAAACA-CAACACCAAUUCAACAAUC---AUGAAAUUCGUAAGUUCAUUGAU .......(((...(((((.(((............))).)))))..)))......(((((....)))))......-................(((---(((((...(....))))).)))) ( -15.60) >consensus AUAUAAGCUGGCCAGCUGGGUUAACUCAACAUCAAACCAAGCUCACAGCUCCAAGUAGUCACUACUACCAAACA_AAACACC_AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAU .....(((((...(((((((((............)))))))))..)))))....((((......)))).................................................... (-14.98 = -16.18 + 1.20)

| Location | 8,176,750 – 8,176,868 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.44 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
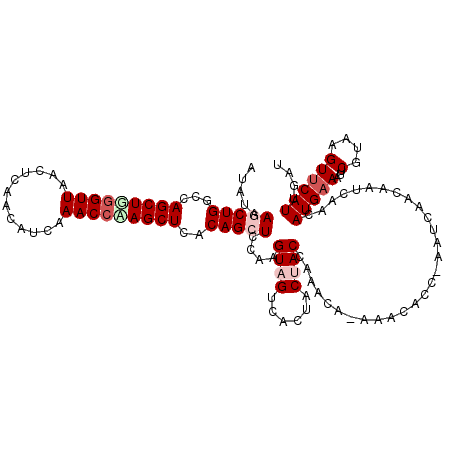
>3L_DroMel_CAF1 8176750 118 + 23771897 GCUCACAGCUCCCAGUAGUCACUUCUACCAAACA-AAGCACC-AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAUUAUCUUAAGGAUUGAUGGGAUCUAUUAAUUAAGGAGUACU ((((..((.(((((((((......))))......-......(-((((...(((((..(((((...(....)))))))))))........))))).))))).))..........))))... ( -20.00) >DroSec_CAF1 4531 112 + 1 GCUCACAGCUCCAAGUAGUCACUACAACCAAACACAAAAACC-AAU-------CAACAUGAAAUUCGUAAGUUCAUUGAUUAUCUUAAGGAUUUAAAGAACUUACCCAUUAAGGAUUACU ((.....))....(((((((.((...................-..(-------(.....)).....((((((((.((((..(((.....))))))).))))))))......))))))))) ( -18.30) >DroSim_CAF1 4506 119 + 1 GCUUACAGCUCCAAGUAGUCACUACAACCAAACACAAAAACC-AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAUUAUCUUAAGGAUUUAAAGGACUUACCCAUUAAGGAUUACU ((.....))....(((((((.((...................-.........((.....)).....((((((((.((((..(((.....))))))).))))))))......))))))))) ( -17.80) >DroEre_CAF1 4734 115 + 1 GCUCACAGCUCCAAGUAGUCACUGCUACCAAACA-AAACCCC-AUUCAACAAUC---AUGAAAUUCGUAAGUCCAUUGAUCACCUAAAGGAUUUAAAGGACUUAAUUAUUAAGGAGUACU .......(((((..(((((....)))))......-.......-...........---....(((...(((((((.((((...((....))..)))).)))))))...)))..)))))... ( -23.90) >DroYak_CAF1 4615 116 + 1 GCUCACAGCUUCAAGUAGUCACUACUACCAAACA-CAACACCAAUUCAACAAUC---AUGAAAUUCGUAAGUUCAUUGAUUACCUUAAGGAUUUAAAGGACUUUCUUAUUAAUUAGCACU .......(((....(((((....)))))......-.....((........((((---(((((...(....))))).))))).((....)).......))...............)))... ( -13.60) >consensus GCUCACAGCUCCAAGUAGUCACUACUACCAAACA_AAACACC_AAUCAACAAUCAACAUGAAAUUCGUAAGUUCAUUGAUUAUCUUAAGGAUUUAAAGGACUUACUCAUUAAGGAGUACU .......(((((..((((......))))......................................((((((((.((((...((....))..)))).)))))))).......)))))... (-13.24 = -14.44 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:35 2006