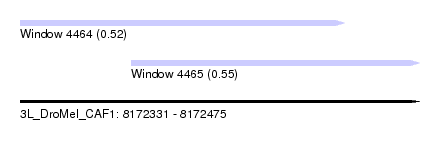
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,172,331 – 8,172,475 |
| Length | 144 |
| Max. P | 0.548992 |

| Location | 8,172,331 – 8,172,448 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -15.03 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
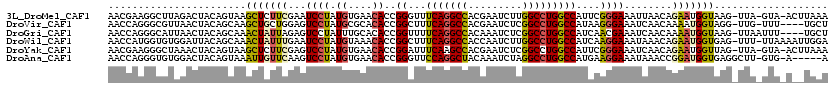
>3L_DroMel_CAF1 8172331 117 + 23771897 AACGAAGGCUUAGACUACAGUAAGCUCUUCGAAUCCUAUGUGAACACCGGGUUUCAGGCCACGAAUCUUGGCCUGGCCAUUCGGGAAAUUAACAGAAUGGUAAG-UUA-GUA-ACUUAAA ..((((((((((........))))).)))))(((((..((....))..))))).(((((((.......)))))))(((((((.(........).)))))))(((-(..-...-))))... ( -38.00) >DroVir_CAF1 164 114 + 1 AACCAGGGCGUUAACUACAGCAAGCUGCUGGAGUCCUAUGCGCACACCGGCUUUCAGGCCACGAAUCUCGGCCUGGCCAUAAGGGAAAUCAACAAAAUGGUAGG-UUG-UUU----UGCU ((((.(.((((..(((.((((.....)))).)))...)))).)..((((((...((((((.........)))))))))....((....))........))).))-)).-...----.... ( -34.30) >DroGri_CAF1 96 115 + 1 AACCAGGGCAUUAACUACAGCAAACUAUUAGAGUCCUAUUUGCACACCGGUUUUCAGGCCACAAAUCUCGGCCUGGCCAUCAACGAAAUCAACAAAAUGGUAAG-UUAAUUU----UGCU ......((((((((((...(((((....(((....))))))))..((((((...((((((.........)))))))))......(....)........))).))-))))...----)))) ( -25.50) >DroWil_CAF1 155 118 + 1 AACCAUGGUGUGGAUUACAGCAAACUAUUUGAAUCCUAUGUAAACACCGGCUUUCAGGCCACCAAUCUUGGCCUGGCCAUCAAGGAAAUAAACAGAAUGGUGAG-UUU-UUAAAAUUGGA .(((((((((((((((.(((........)))))))).......)))))(((...(((((((.......))))))))))..................)))))...-...-........... ( -31.21) >DroYak_CAF1 94 117 + 1 AACGAAGGGCUAAACUACAGUAAGCUCUUCGAGUCCUAUGUGAACACCGGAUUUCAAGCCACGAAUCUCGGCCUGGCCAUUCGGGAAAUCAACAGAAUGGUUAG-UUA-GUA-ACUUAAA ..(((((((((..((....)).)))))))))(((((..((....))..)))))....(((.........)))((((((((((.(........).))))))))))-...-...-....... ( -34.70) >DroAna_CAF1 94 113 + 1 AACCAGGGUGUGGACUACAGUAAAUUGUUCAAGUCCUAUGUGAACACCGGGUUCCAGGCUACAAAUCUAGGCCUGGCCAUGAAGGAAAUAAACCGGAUGGUGAGGCUU-GUG-A-----A ((((..((((((((((((((....))))...))))).......))))).))))............(((((((((.(((((...((.......))..))))).))))))-).)-)-----. ( -35.11) >consensus AACCAGGGCGUAAACUACAGCAAACUAUUCGAGUCCUAUGUGAACACCGGCUUUCAGGCCACGAAUCUCGGCCUGGCCAUCAAGGAAAUCAACAGAAUGGUAAG_UUA_GUA_A__UGAA .......................(((((((...((((.((....))..((...(((((((.........)))))))))....))))........)))))))................... (-15.03 = -15.87 + 0.84)

| Location | 8,172,371 – 8,172,475 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -18.50 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8172371 104 + 23771897 UGAACACCGGGUUUCAGGCCACGAAUCUUGGCCUGGCCAUUCGGGAAAUUAACAGAAUGGUAAGUUAGUAACUUAAAUACAUA--ACUACGAUAUGUUACAAACCC ........(((((((((((((.......)))))))(((((((.(........).))))))).((((((((.......))).))--)))............)))))) ( -30.30) >DroVir_CAF1 204 99 + 1 CGCACACCGGCUUUCAGGCCACGAAUCUCGGCCUGGCCAUAAGGGAAAUCAACAAAAUGGUAGGUUGUUU---UGCUUUCUGGAAACCACACU-AC--ACA-AUAC ........((.(((((((...((((...(((((((.((((...(........)...))))))))))).))---))...))))))).)).....-..--...-.... ( -23.70) >DroSec_CAF1 134 104 + 1 UGACCACCGGGUUUCAGGCCACGAAUCUUGGGCUGGCCAUUCGUGAAAUCAACAGAAUGGUAAGUUAGUAACUUAAAUACGAA--ACUCCGAUAUGUUACAAACCC ........(((((((((.(((.......))).)))(((((((.((....))...)))))))......(((((....(((((..--....)).)))))))))))))) ( -27.20) >DroSim_CAF1 134 104 + 1 UGACCACCGGGUUUCAGGCCACGAAUCUUGGGCUGGCCAUUCGUGAAAUCAACAGAAUGGUAAGUUAGUAACUUUAAUACGAA--ACUCCGAUAUGUUACAAACCC ........(((((((((.(((.......))).)))(((((((.((....))...)))))))......(((((....(((((..--....)).)))))))))))))) ( -27.20) >DroEre_CAF1 134 104 + 1 UGAACACCGGGUUUCAGGCCACGAAUCUUGGCCUGGCCAUUCGGGAGAUCAACAGAAUGGUUAGUUAGUAACUUAAAUACUCA--AGUCCGAUAUGUUACAAACCC ........(((((((((((((.......)))))))(((((((.(........).)))))))......(((((......((...--.)).......))))))))))) ( -30.02) >DroYak_CAF1 134 104 + 1 UGAACACCGGAUUUCAAGCCACGAAUCUCGGCCUGGCCAUUCGGGAAAUCAACAGAAUGGUUAGUUAGUAACUUAAAUAUUCA--ACUCCGAUAUGUUACAAGUCC ........((((((...(((.........)))((((((((((.(........).))))))))))...(((((....(((((..--.....)))))))))))))))) ( -23.20) >consensus UGAACACCGGGUUUCAGGCCACGAAUCUUGGCCUGGCCAUUCGGGAAAUCAACAGAAUGGUAAGUUAGUAACUUAAAUACUAA__ACUCCGAUAUGUUACAAACCC ........((((((((((((.........))))))(((((((............)))))))......(((((.......................))))))))))) (-18.50 = -20.02 + 1.51)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:31 2006