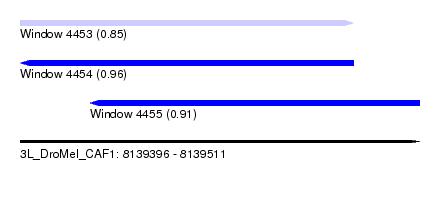
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,139,396 – 8,139,511 |
| Length | 115 |
| Max. P | 0.955826 |

| Location | 8,139,396 – 8,139,492 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
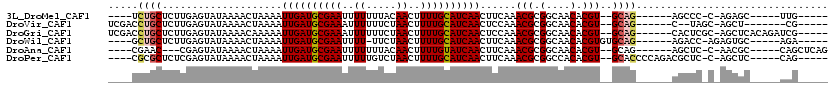
>3L_DroMel_CAF1 8139396 96 + 23771897 -----CAA-----GCUCU-G-GGGCU------CUGC--ACGUGUUGCCGCGUUUGAAGUUGAUGCAAAAGUUGUAAAAAAAUUCGCAUCAAUUUUAGUUUUAUACUCAAGAGCAGA---- -----...-----(((((-(-((...------..((--(((((....))))).((((((((((((.((..((.....))..)).))))))))))))))......))).)))))...---- ( -28.10) >DroVir_CAF1 36741 97 + 1 -----CG-------AGCU-GCUA--G------CUGC--ACGUGUUGCCGCGUUUGGAGUUGAUGCAAAAGUUAGAAAAAAUUUCGCAUCAAUUUUAGUUUUAUACUCAAGAGCAGGUCGA -----((-------(.((-(((.--(------(.((--(.....))).))..(((((((((((((..(((((......))))).))))))))))))).............))))).))). ( -31.10) >DroGri_CAF1 37186 106 + 1 -----CGAUCUGUGAGCU-GCGAGUG------CUGC--ACGUGUUGCCGCGUUUGGAGUUGAUGCAAAAGUUAGAAAAAAUUUCGCAUCAAUUUUUGUUUUAUACUCAAGAGCAGGUCGA -----((((((((...((-..(((((------..((--(((((....)))))..(((((((((((..(((((......))))).))))))))))).))....))))).)).)))))))). ( -33.80) >DroWil_CAF1 44699 98 + 1 -----UCU-----GCACUCU-GGUCU------CUGCACACGUGUUGCCGCGUUUGAAGUUGAUGCAAAAGUUAGAA-AAAAUUCGCAUCAAUUUUAGUUUUAUACUCAAGAGCAGC---- -----.((-----((.((..-(((..------..((..(((((....))))).((((((((((((...((((....-..)))).)))))))))))))).....)))..)).)))).---- ( -24.40) >DroAna_CAF1 72279 98 + 1 CUGAGCUG-----GCGUU-G-GAGCU------CUGC--ACGUGUUGCCGCGUUUGAAGUUGAUACAAAAGUUGUAAAAAAAUUCGCAUCAAUUUUAGUUUUAUACUCG---GUUCG---- ..((((((-----(.(((-(-((((.------....--(((((....))))).((((((((((.(.((..((.....))..)).).)))))))))))))))).)))))---)))).---- ( -22.40) >DroPer_CAF1 39096 102 + 1 -----CUG-----GAGCU-G-GAGCGUCUGGGGUGC--ACGUGUGGCCGCGUUUGAAGUUGAUGCAAAAGUUAGACAAAAAUUCGCAUCAAUUUUAGUUUUAUACUCGAGAGCGCG---- -----...-----.....-.-..((((((.(((((.--(((((....)))))(((((((((((((...................))))))))))))).....))))).))).))).---- ( -30.31) >consensus _____CGA_____GAGCU_G_GAGCG______CUGC__ACGUGUUGCCGCGUUUGAAGUUGAUGCAAAAGUUAGAAAAAAAUUCGCAUCAAUUUUAGUUUUAUACUCAAGAGCAGG____ ................................((((..(((((....)))))(((((((((((((...................)))))))))))))..............))))..... (-18.10 = -18.08 + -0.03)


| Location | 8,139,396 – 8,139,492 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8139396 96 - 23771897 ----UCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUUUACAACUUUUGCAUCAACUUCAAACGCGGCAACACGU--GCAG------AGCCC-C-AGAGC-----UUG----- ----...(((((.(.((............((((((((((.............))))))))))..((..(((.(....).)))--...)------))).)-.-)))))-----...----- ( -23.72) >DroVir_CAF1 36741 97 - 1 UCGACCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAAUUUUUUCUAACUUUUGCAUCAACUCCAAACGCGGCAACACGU--GCAG------C--UAGC-AGCU-------CG----- .(((.(((((....(((............(((((((((((.((.....)).)))))))))))......(((.(....).)))--...)------)--))))-)).)-------))----- ( -25.40) >DroGri_CAF1 37186 106 - 1 UCGACCUGCUCUUGAGUAUAAAACAAAAAUUGAUGCGAAAUUUUUUCUAACUUUUGCAUCAACUCCAAACGCGGCAACACGU--GCAG------CACUCGC-AGCUCACAGAUCG----- ..((.((((....((((............(((((((((((.((.....)).)))))))))))........((.(((.....)--)).)------)))))))-)).))........----- ( -26.30) >DroWil_CAF1 44699 98 - 1 ----GCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUU-UUCUAACUUUUGCAUCAACUUCAAACGCGGCAACACGUGUGCAG------AGACC-AGAGUGC-----AGA----- ----.((((.(((..((............((((((((((..((-....))..))))))))))(((...(((((......)))))...)------)))).-.))).))-----)).----- ( -28.70) >DroAna_CAF1 72279 98 - 1 ----CGAAC---CGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUUUACAACUUUUGUAUCAACUUCAAACGCGGCAACACGU--GCAG------AGCUC-C-AACGC-----CAGCUCAG ----.(((.---..(((.....)))....((((((((((.............)))))))))).)))..(((.(....).)))--...(------((((.-.-.....-----.))))).. ( -20.02) >DroPer_CAF1 39096 102 - 1 ----CGCGCUCUCGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUGUCUAACUUUUGCAUCAACUUCAAACGCGGCCACACGU--GCACCCCAGACGCUC-C-AGCUC-----CAG----- ----.(((.(((.(.((............((((((((((..((.....))..)))))))))).......((((......)))--).)).).))))))..-.-.....-----...----- ( -21.70) >consensus ____CCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUUUCUAACUUUUGCAUCAACUUCAAACGCGGCAACACGU__GCAG______AGCUC_C_AGCGC_____CAG_____ .....((((....................((((((((((..((.....))..))))))))))......(((.(....).)))..))))................................ (-15.48 = -15.78 + 0.31)

| Location | 8,139,416 – 8,139,511 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
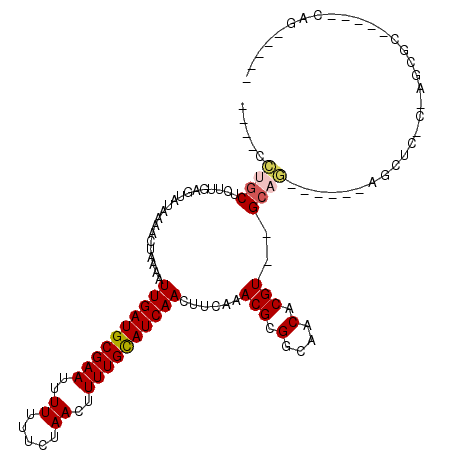
>3L_DroMel_CAF1 8139416 95 - 23771897 GAGUCCA---ACG----AGCC-----AGCGC----UCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUUUACAACUUUUGCAUCAACUUCAAACGCGGCAACAC .......---...----.(((-----.((((----((.......))))............((((((((((.............))))))))))........)))))..... ( -21.82) >DroVir_CAF1 36758 97 - 1 -UGGUA----ACG----GCGC-----CGCGCUCGACCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAAUUUUUUCUAACUUUUGCAUCAACUCCAAACGCGGCAACAC -.....----...----..((-----(((((((((.......))))))............(((((((((((.((.....)).)))))))))))........)))))..... ( -27.60) >DroPse_CAF1 46005 98 - 1 GACCCCG---GAC----AACCCCG--AGCAA----CGCGCUCUCGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUGUCUAACUUUUGCAUCAACUUCAAACGCGGCCACAC .....((---(..----....)))--.....----((((......(((.....)))....((((((((((..((.....))..)))))))))).......))))....... ( -19.90) >DroWil_CAF1 44722 103 - 1 GACCAAG---CGGCGGCAACAUCGGCGGCUG----GCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUU-UUCUAACUUUUGCAUCAACUUCAAACGCGGCAACAC .......---((((.((.......)).))))----(((((..((((((.....)).....((((((((((..((-....))..))))))))))..))))..)))))..... ( -29.20) >DroMoj_CAF1 37695 102 - 1 GUGGCAAACGACG----CCGC-----CGCGCUCGACCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAAUUUUUUCUAACUUUUGCAUCAACUCCAAACGCGGCAACAC ..(((.......)----))((-----(((((((((.......))))))............(((((((((((.((.....)).)))))))))))........)))))..... ( -31.60) >DroPer_CAF1 39122 98 - 1 GACCCCG---GAC----AACCCCG--AGCAA----CGCGCUCUCGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUGUCUAACUUUUGCAUCAACUUCAAACGCGGCCACAC .....((---(..----....)))--.....----((((......(((.....)))....((((((((((..((.....))..)))))))))).......))))....... ( -19.90) >consensus GACCCAG___ACG____AACC_____AGCGC____CCUGCUCUUGAGUAUAAAACUAAAAUUGAUGCGAAUUUUUUUCUAACUUUUGCAUCAACUUCAAACGCGGCAACAC ...........................((.........(((....)))............((((((((((..((.....))..))))))))))........))(....).. (-13.87 = -13.87 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:20 2006