| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 816,004 – 816,112 |
| Length | 108 |
| Max. P | 0.814563 |

| Location | 816,004 – 816,112 |
|---|---|
| Length | 108 |
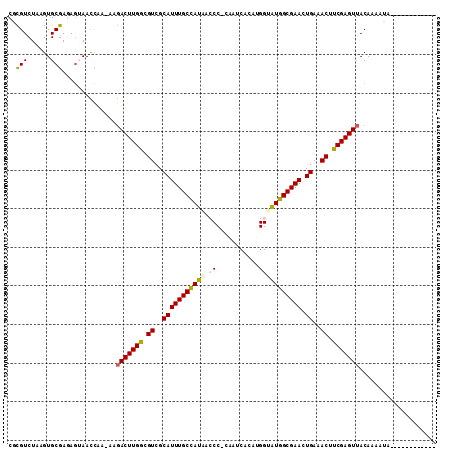
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
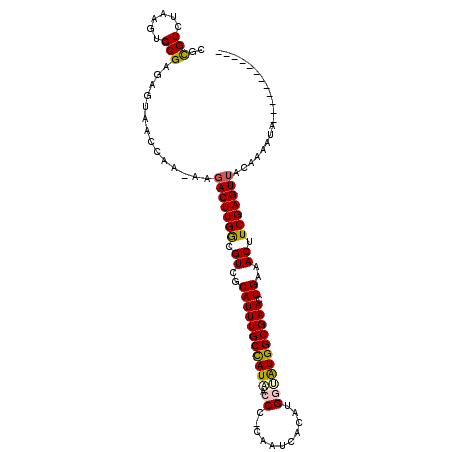
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 816004 108 - 23771897 CGCGUCUAAGUGCGAGAGAAACCGA-AAGACUUGACGUCGCAUUUGCCAUAAACC-AACUCA-ACGGAGUGGCGAAUUGAAACUUCGAGUUACAAAGUCAAUAAAAUAUCA ((((......))))...........-..(((((((.((..((((((((((...((-......-..)).)))))))).))..)).))))))).................... ( -24.30) >DroVir_CAF1 199983 94 - 1 CGUGUCUAAGUGCGAUAUUAAUCC----AACUUGGCGUCGCAUUUGCCAUAACCC-CAAUCACAUGGUAUGGCGAACUGAAACUUCGAGUAACAAGAUA------------ ..(((((...((.(((....))))----)((((((.((..(((((((((((...(-((......)))))))))))).))..)).))))))....)))))------------ ( -23.60) >DroPse_CAF1 186879 98 - 1 CCCGUCUACGUGCGAGAGUAACCAAAAAGACUUGGCGUCGCAUUUGCCAUAACCC-CAAACACAUGGUAUGGCGAACUGAAACUUCGAGUUACACCCGA------------ ........(((((....)))........(((((((.((..(((((((((((...(-((......)))))))))))).))..)).))))))).....)).------------ ( -24.80) >DroEre_CAF1 179253 106 - 1 CACGUCUAAGUGCGAGAGAAACCGA-AAGACUUGGCGUCGCAUUUGCCAUAAACC-AGCUCA-ACGGAAUGGCGAAUUGAAACUUCGAGUUACAAAGUCAAUAAAAUG--A (((......))).............-..(((((((.((..((((((((((...((-......-..)).)))))))).))..)).))))))).................--. ( -21.30) >DroMoj_CAF1 196223 95 - 1 CGUGUCUAUGUGCGAUAGUAAUUU----GACUUGACGUCGCAUUUGCUAUGACCCUUAACCACAUGGCAUGGCGAACUGAAACUUCGAGUUACAAGAAC------------ ....(((...(((....)))...(----(((((((.((..(((((((((((.((...........))))))))))).))..)).))))))))..)))..------------ ( -24.00) >DroPer_CAF1 193029 98 - 1 CCCGUCUACGUGCGAGAGUAACCAAAAAGACUUGGCGUCGCAUUUGCCAUAACCC-CAAACACGUGGUAUGGCGAACUGAAACUUCGAGUUACACCCGA------------ ........(((((....)))........(((((((.((..(((((((((((.((.-(......).))))))))))).))..)).))))))).....)).------------ ( -25.50) >consensus CGCGUCUAAGUGCGAGAGUAACCAA_AAGACUUGGCGUCGCAUUUGCCAUAACCC_CAAUCACAUGGUAUGGCGAACUGAAACUUCGAGUUACAAAAUA____________ ..(((......)))..............(((((((.((..(((((((((((.((...........))))))))))).))..)).))))))).................... (-19.17 = -19.12 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:19 2006