
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,127,360 – 8,127,452 |
| Length | 92 |
| Max. P | 0.999953 |

| Location | 8,127,360 – 8,127,452 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 65.51 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -14.48 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
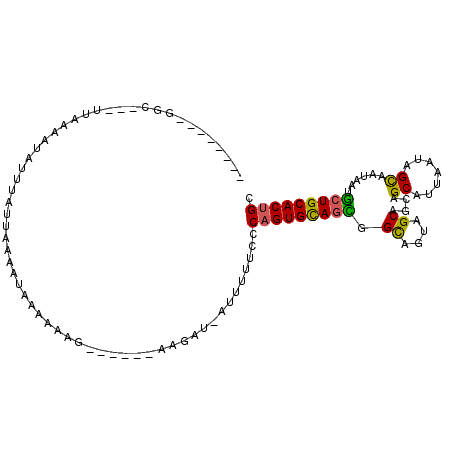
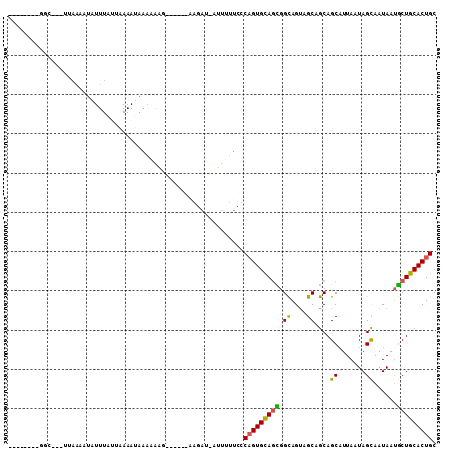
>3L_DroMel_CAF1 8127360 92 + 23771897 --------GGC---UUAAAAUAUUUAGUAAAAUAAAAAAG------AAGAUCAUUCUUCUCAGUGCAGCGGCAGUAGCAGCAGCAUUACUAGCAAUAAUGCUGCACUGC --------...---........................((------((((....))))))((((((((((((((((((....))..)))).)).....)))))))))). ( -25.50) >DroSec_CAF1 24095 91 + 1 --------G-G---UUAAAAUAUUUAUUAAAAUGAACAAG------AAGAUAAUUGUUCUCAGUGCAGCGGCAGUAGCAGCAGCAUUAAUAGCAAUAAUGCUGCACUGC --------.-.---...................((((((.------.......)))))).((((((.((.((....)).))(((((((.......))))))))))))). ( -24.70) >DroYak_CAF1 24393 100 + 1 AUACUUAAGGCUUCUAAAAAUAUUU--UCAAAGGAAAAAAUAUAUU-------UUUUUUCCUGUGCAGUGGCAGUAGCAGCAGCCUUAAUAGCAAUAAUGCUGCACUGC ....((((((((.((..........--....(((((((((......-------))))))))).(((....))).....)).))))))))(((((....)))))...... ( -25.00) >DroAna_CAF1 22898 91 + 1 ---------GC---UUCCUGUUUUUUUUCAAAUAUUAGUU------UGGAUUAUUUCUCCCAGUGUACUAGUAGCUGCCACAGCAUUAAUAGUAAUAACACUGCACUGC ---------..---............(((((((....)))------))))..........((((((((((...((((...)))).....))))....))))))...... ( -14.70) >consensus ________GGC___UUAAAAUAUUUAUUAAAAUAAAAAAG______AAGAU_AUUUUUCCCAGUGCAGCGGCAGUAGCAGCAGCAUUAAUAGCAAUAAUGCUGCACUGC ............................................................(((((((((.((....))....((.......))......))))))))). (-14.48 = -13.97 + -0.50)

| Location | 8,127,360 – 8,127,452 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 65.51 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.81 |
| SVM RNA-class probability | 0.999953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
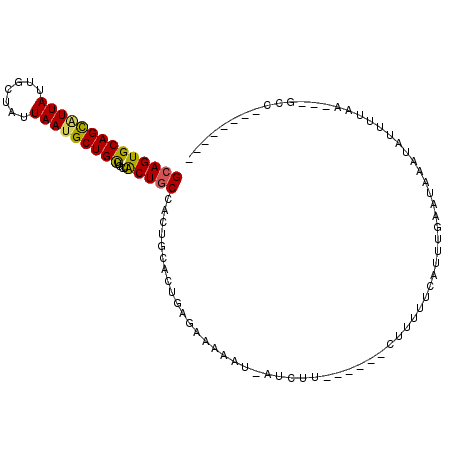
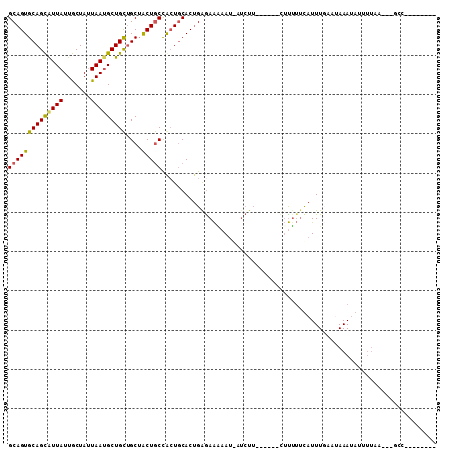
>3L_DroMel_CAF1 8127360 92 - 23771897 GCAGUGCAGCAUUAUUGCUAGUAAUGCUGCUGCUACUGCCGCUGCACUGAGAAGAAUGAUCUU------CUUUUUUAUUUUACUAAAUAUUUUAA---GCC-------- (((((((((((.(((((....))))).)))))).))))).(((.....(((((((....))))------)))...(((((....))))).....)---)).-------- ( -28.10) >DroSec_CAF1 24095 91 - 1 GCAGUGCAGCAUUAUUGCUAUUAAUGCUGCUGCUACUGCCGCUGCACUGAGAACAAUUAUCUU------CUUGUUCAUUUUAAUAAAUAUUUUAA---C-C-------- ((((((((((((((.......))))))))).((....)).))))).....((((((.......------.))))))...................---.-.-------- ( -26.20) >DroYak_CAF1 24393 100 - 1 GCAGUGCAGCAUUAUUGCUAUUAAGGCUGCUGCUACUGCCACUGCACAGGAAAAAA-------AAUAUAUUUUUUCCUUUGA--AAAUAUUUUUAGAAGCCUUAAGUAU (((((((((((.....(((.....))))))))).))))).....((.(((((((((-------......))))))))).)).--..((((((..((....)).)))))) ( -27.20) >DroAna_CAF1 22898 91 - 1 GCAGUGCAGUGUUAUUACUAUUAAUGCUGUGGCAGCUACUAGUACACUGGGAGAAAUAAUCCA------AACUAAUAUUUGAAAAAAAACAGGAA---GC--------- ((...(((((((((.......))))))))).)).(((.((.((......(((.......))).------........(((....))).)).)).)---))--------- ( -16.50) >consensus GCAGUGCAGCAUUAUUGCUAUUAAUGCUGCUGCUACUGCCACUGCACUGAGAAAAAU_AUCUU______CUUUUUCAUUUGAAUAAAUAUUUUAA___GCC________ ((((((((((((((.......)))))))))....)))))...................................................................... (-15.06 = -14.88 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:16 2006