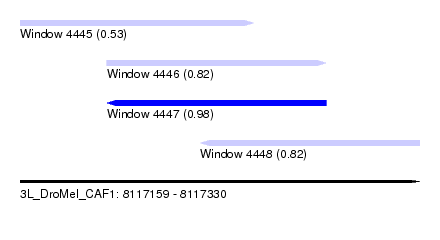
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,117,159 – 8,117,330 |
| Length | 171 |
| Max. P | 0.979832 |

| Location | 8,117,159 – 8,117,259 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -46.87 |
| Consensus MFE | -26.27 |
| Energy contribution | -27.68 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533488 |
| Prediction | RNA |
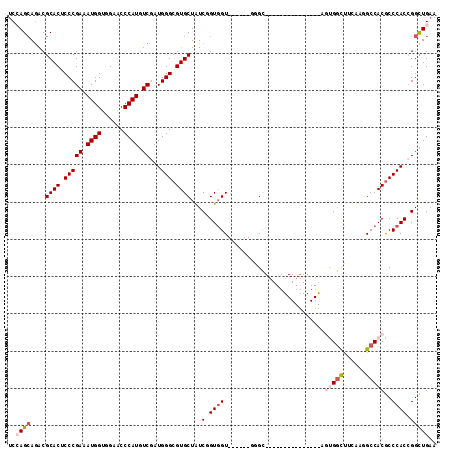
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8117159 100 + 23771897 GCUAUCAGGA---UUACCCCUCAACAGAGGAGCCCCAUGCUCGAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCGAUGGGCGUGCUAUCGGUGGU------UGGC----------- (((.......---.....((((....))))((((((.(((....)))...((((.(((((.((((......)))).))...))).))))....)).)))------))))----------- ( -31.20) >DroVir_CAF1 14411 114 + 1 GCUGCUCGGCUGUUCGCCGCGCAGCAGCGGCGCUCCAUGCUCCAGCAGGCGCACGCCCGAUAUGGUUGAGCCCAUGUCGAUGGGCGUGCUGUCUGUGGU------GCGCGUAUUGCUGGU (((((.((((.....)))).)))))(((.((((.....((.(((.((((((((((((((((((((......)))))))...)))))))).)))))))).------))))))...)))... ( -63.70) >DroPse_CAF1 22662 106 + 1 GCUGUCGGGC---CGUCCUCGCAGCAGCGGUGCCCCGUGCUCCAGCAAACGCACCCCCGAGAUGGUCGAACCCAUGUCGAUGGGCGUGCUGUCUGUCGUCACUGGCGGC----------- (((((((((.---((.(...(((((((((((((....(((....)))...)))))(((((.((((......)))).)))..)))).))))))..).)).).))))))))----------- ( -45.40) >DroYak_CAF1 14086 100 + 1 GCUAUCGGGA---CUUCCUCGCAACAGAGGAGCCCCAUGCUCGAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCAAUGGGCGUGCUAUCGGUGGU------UGGC----------- (((((((((.---((.((((......)))))).))).(((....)))...((((.(((((.((((......)))).))...))).))))....))))))------....----------- ( -36.20) >DroMoj_CAF1 13992 114 + 1 GCUGUCUGGCUGUUCGCCGCGCAGCAGUGGAGCUCCAUGCUCCAGCAGGCGCACGCCCGAUAUGGUCGAGCCCAUGUCAAUGGGCGUGCUGUCUGUGGU------GCGCGCCUCAUUGGC .......(((.....)))(((((.(..((((((.....))))))(((((((((((((((((((((......)))))))...)))))))).))))))).)------))))(((.....))) ( -62.80) >DroAna_CAF1 14053 100 + 1 GCUAUCCGGA---UCGCCUCUUAGCAAGGGGGCGCCAUGCUCCAGGAGACGCACUCCCGAAAUGGUGGAGCCCAUGUCAAUGGGCGUGCUAUCGGUGGU------GGGG----------- ....((((((---.((((((((....))))))))......))).)))..(.(((..((((..(((((..((((((....)))))).)))))))))..))------).).----------- ( -41.90) >consensus GCUAUCCGGA___UCGCCUCGCAGCAGAGGAGCCCCAUGCUCCAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCAAUGGGCGUGCUAUCGGUGGU______GGGC___________ (((((.(((.......))).)))))...(((((.....))))).......((((.(((((.((((......)))).))...))).))))............................... (-26.27 = -27.68 + 1.42)

| Location | 8,117,196 – 8,117,290 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -22.39 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
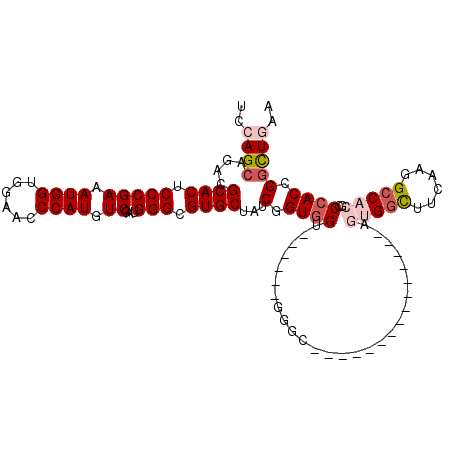
>3L_DroMel_CAF1 8117196 94 + 23771897 UCGAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCGAUGGGCGUGCUAUCGGUGGU------UGGC---------------AAUGGUUUUAAGGCCACACCCACCGGCUGAA ((.(((....((((.(((((.((((......)))).))...))).))))...((((((.------((..---------------..(((((....))))))).))))))))))). ( -34.00) >DroVir_CAF1 14451 109 + 1 UCCAGCAGGCGCACGCCCGAUAUGGUUGAGCCCAUGUCGAUGGGCGUGCUGUCUGUGGU------GCGCGUAUUGCUGGUCAGUUGCGGCCGCACUGCCACGCCCACCGGCUGAA ..(((((((((((((((((((((((......)))))))...)))))))).))))((((.------(((.(((.(((.((((......))))))).)))..)))))))..)))).. ( -55.90) >DroPse_CAF1 22699 100 + 1 UCCAGCAAACGCACCCCCGAGAUGGUCGAACCCAUGUCGAUGGGCGUGCUGUCUGUCGUCACUGGCGGC---------------ACUGGCUUCAGGGCCACGCCCACCGGCUGAA ..((((...........(((.((((......)))).))).(((((((((((((.((....)).))))))---------------))(((((....))))).)))))...)))).. ( -43.60) >DroGri_CAF1 14128 109 + 1 UCCAGCAGGCGCACACCCGAUAUGGUCGAGCCCAUGUCGAUGGGCGUGCUGUCUGUGGU------GCGUGCAUCGCUGCUCAGCUGCGGCCUCACUGCCACGCCCACCGGCUGCA ..((((((((((((.((((((((((......)))))))...))).)))).))))((((.------(((((((..(((((......))))).....)).)))))))))..)))).. ( -51.20) >DroYak_CAF1 14123 94 + 1 UCGAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCAAUGGGCGUGCUAUCGGUGGU------UGGC---------------AGUGGUUUUAGGGCCACGCCCACCGGUUGAA ((((.(....((((.(((((.((((......)))).))...))).))))....(((((.------..((---------------.((((((....)))))))))))))).)))). ( -36.50) >DroAna_CAF1 14090 94 + 1 UCCAGGAGACGCACUCCCGAAAUGGUGGAGCCCAUGUCAAUGGGCGUGCUAUCGGUGGU------GGGG---------------AGUGAUUUUAAUGCCACGCCCACGGGCUGAA ....(....).(((((((.(..((((((.((((((....))))))...))))))....)------.)))---------------))))..........((.(((....))))).. ( -38.90) >consensus UCCAGCAGACGCACUCCCGAAAUGGUGGAACCCAUGUCGAUGGGCGUGCUAUCGGUGGU______GGGC_______________AGUGGCUUCAAGGCCACGCCCACCGGCUGAA ..((((....((((.(((((.((((......)))).))...))).))))...(.((((...........................(((((......)))))..)))).))))).. (-22.39 = -23.50 + 1.11)
| Location | 8,117,196 – 8,117,290 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
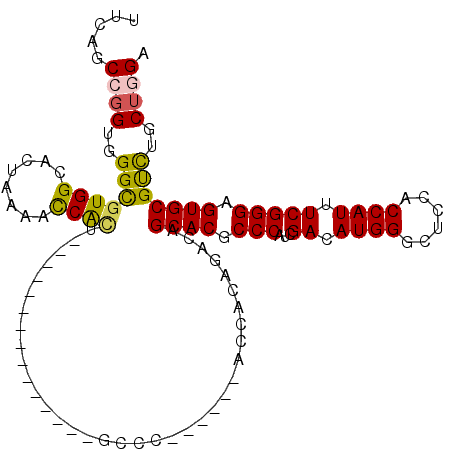
>3L_DroMel_CAF1 8117196 94 - 23771897 UUCAGCCGGUGGGUGUGGCCUUAAAACCAUU---------------GCCA------ACCACCGAUAGCACGCCCAUCGACAUGGGUUCCACCAUUUCGGGAGUGCGUCUGCUCGA .(((((((((((...((((............---------------))))------.))))))...((((.(((...((.((((......)))).))))).))))....))).)) ( -35.80) >DroVir_CAF1 14451 109 - 1 UUCAGCCGGUGGGCGUGGCAGUGCGGCCGCAACUGACCAGCAAUACGCGC------ACCACAGACAGCACGCCCAUCGACAUGGGCUCAACCAUAUCGGGCGUGCGCCUGCUGGA .....((((..(((((((..(((((...((.........))....)))))------.)))).....((((((((...((.((((......)))).)))))))))))))..)))). ( -53.70) >DroPse_CAF1 22699 100 - 1 UUCAGCCGGUGGGCGUGGCCCUGAAGCCAGU---------------GCCGCCAGUGACGACAGACAGCACGCCCAUCGACAUGGGUUCGACCAUCUCGGGGGUGCGUUUGCUGGA .....((((..((((((((.(((....))).---------------)))))...............((((.(((..(((.((((......)))).)))))))))))))..)))). ( -43.20) >DroGri_CAF1 14128 109 - 1 UGCAGCCGGUGGGCGUGGCAGUGAGGCCGCAGCUGAGCAGCGAUGCACGC------ACCACAGACAGCACGCCCAUCGACAUGGGCUCGACCAUAUCGGGUGUGCGCCUGCUGGA .....((((..(((((((..((..(((....)))..)).(((.....)))------.)))).....((((((((...((.((((......)))).)))))))))))))..)))). ( -51.40) >DroYak_CAF1 14123 94 - 1 UUCAACCGGUGGGCGUGGCCCUAAAACCACU---------------GCCA------ACCACCGAUAGCACGCCCAUUGACAUGGGUUCCACCAUUUCGGGAGUGCGUCUGCUCGA ......((((((((((((........)))).---------------))..------.))))))...((((.(((...((.((((......)))).))))).)))).......... ( -33.90) >DroAna_CAF1 14090 94 - 1 UUCAGCCCGUGGGCGUGGCAUUAAAAUCACU---------------CCCC------ACCACCGAUAGCACGCCCAUUGACAUGGGCUCCACCAUUUCGGGAGUGCGUCUCCUGGA (((((...(((((.((((........)))).---------------.)))------))....(((.((((.(((...((.((((......)))).))))).)))))))..))))) ( -31.30) >consensus UUCAGCCGGUGGGCGUGGCACUAAAACCACU_______________GCCC______ACCACAGACAGCACGCCCAUCGACAUGGGCUCCACCAUUUCGGGAGUGCGUCUGCUGGA .....((((..(((((((........))))....................................((((.(((...((.((((......)))).))))).)))))))..)))). (-32.50 = -32.20 + -0.30)
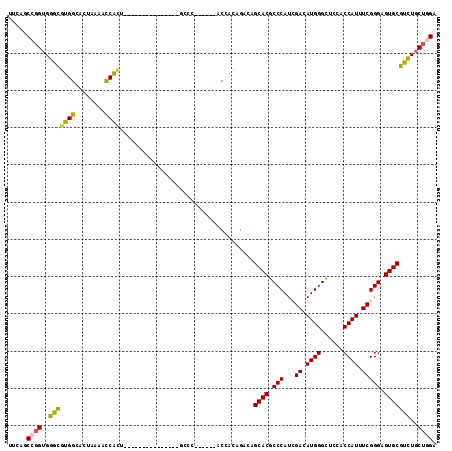
| Location | 8,117,236 – 8,117,330 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.19 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8117236 94 - 23771897 CUACCAUCCAGGGGCGGCAGUCCAUAUCUGCCCCGUGGCCUUCAGCCGGUGGGUGUGGCCUUAAAACCAUU---------------GCCA------ACCACCGAUAGCACGCCCA ...........(((((((((.......)))))..(((((.....))((((((...((((............---------------))))------.))))))....))))))). ( -35.90) >DroVir_CAF1 14491 109 - 1 CUGCCCUCGCGUGGUGGCAGCCCAUAUUUGCCGCGAGGCCUUCAGCCGGUGGGCGUGGCAGUGCGGCCGCAACUGACCAGCAAUACGCGC------ACCACAGACAGCACGCCCA .(((..((..((((((((.((((((....(((....))).........)))))).(((((((((....)).)))).))).......)).)------))))).))..)))...... ( -44.32) >DroPse_CAF1 22739 100 - 1 CUGCCCUCCCGGGGCGGAAGCCCCUAUCUGCCGCGUGGCCUUCAGCCGGUGGGCGUGGCCCUGAAGCCAGU---------------GCCGCCAGUGACGACAGACAGCACGCCCA ..((((.((.(((((....)))))............(((.....))))).))))(((((.(((....))).---------------)))))..(((.(........))))..... ( -43.70) >DroSec_CAF1 14157 94 - 1 CUACCCUCCAGGGGCGGUAGUCCAUACCUGCCCCGUGGCCUUCAGCCGGUGGGUGUGGCCUUAAAACCAUU---------------ACCA------ACCACCGACAGCACGCCCA ..........(((((((((.....)))).)))))(.(((.......((((((..((((........)))).---------------....------.)))))).......)))). ( -33.84) >DroYak_CAF1 14163 94 - 1 CUGCCCUCCAGGGGCGGCAGUCCAUAUCUGCCCCGAGGUCUUCAACCGGUGGGCGUGGCCCUAAAACCACU---------------GCCA------ACCACCGAUAGCACGCCCA .(((((((..(((((((..........)))))))))))........((((((((((((........)))).---------------))..------.))))))...)))...... ( -35.30) >DroAna_CAF1 14130 94 - 1 CUGCCCUCCAGAGGCGGAAGUCCCUAUCUGCCCCGUGGCCUUCAGCCCGUGGGCGUGGCAUUAAAAUCACU---------------CCCC------ACCACCGAUAGCACGCCCA ((((((....).)))))..((..(((((.((((((.(((.....))))).))))((((.............---------------....------.)))).))))).))..... ( -29.37) >consensus CUGCCCUCCAGGGGCGGCAGUCCAUAUCUGCCCCGUGGCCUUCAGCCGGUGGGCGUGGCCUUAAAACCACU_______________GCCA______ACCACCGACAGCACGCCCA ..(((.....(((((((..........)))))))..(((.....))))))(((((((..................................................))))))). (-26.86 = -26.19 + -0.66)
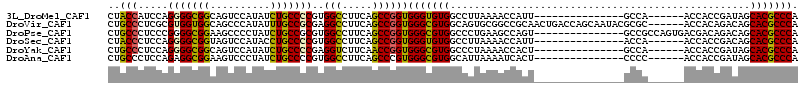
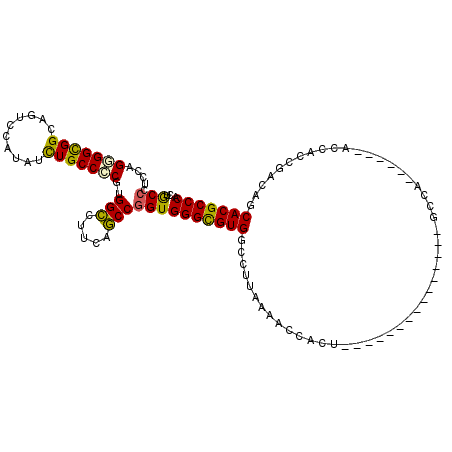

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:14 2006