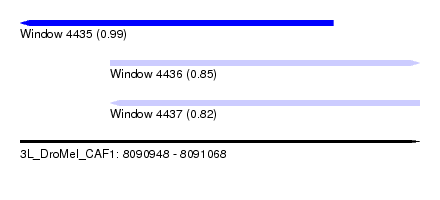
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,090,948 – 8,091,068 |
| Length | 120 |
| Max. P | 0.994910 |

| Location | 8,090,948 – 8,091,042 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8090948 94 - 23771897 GCGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUCUACAGUGGGGCCAAAGCAAUUGGUUAGUGG--GGUGG .(.((((..((((((((((...((((((...((((...))))..)))))))))))))))).)))).)((((((.....)))))).....--..... ( -32.10) >DroSec_CAF1 1963 94 - 1 GCGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUCUACAGUGGGGCCAAAGCAAUUGGUUAGUGG--GGUGG .(.((((..((((((((((...((((((...((((...))))..)))))))))))))))).)))).)((((((.....)))))).....--..... ( -32.10) >DroSim_CAF1 3003 94 - 1 GCGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUCUACAGUGGGGCCAAAGCAAUUGGUUAGUGG--GGUGG .(.((((..((((((((((...((((((...((((...))))..)))))))))))))))).)))).)((((((.....)))))).....--..... ( -32.10) >DroEre_CAF1 1964 94 - 1 UUGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCAUAAUGAGCAUGCCUCUACAGUGGGGCCAAAGCAAAUGGUCAGUGG--GGUGG .(.((((..((((((((((...((((((...((((...))))..)))))))))))))))).)))).)(((((.......))))).....--..... ( -32.50) >DroYak_CAF1 2138 94 - 1 GUGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUCUACAGUGGGGCCAAAUCCAUUGGUCAGUGG--GGUGG .(.(((((.((((((((((...((((((...((((...))))..)))))))))))))))).(((((((......))))))).))))).)--..... ( -33.40) >DroAna_CAF1 2160 95 - 1 GCGA-UGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUGGACAGUGGUGCCGAAGCAAUUGGUUAAUUGCGGCCGG ....-......((((((((...((((((...((((...))))..))))))))))))))((...(..(((((((.....)))))..))..)..)).. ( -28.70) >consensus GCGAUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUCUACAGUGGGGCCAAAGCAAUUGGUUAGUGG__GGUGG .(.((((..((((((((((...((((((...((((...))))..)))))))))))))))).)))).)((((((.....))))))............ (-29.86 = -30.17 + 0.31)


| Location | 8,090,975 – 8,091,068 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -12.98 |
| Energy contribution | -14.98 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8090975 93 + 23771897 CCACUGUA---GAGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCCUCCCAAU-C---GCCUCCCACC-AACCCCCCUGUUGUCAAU ........---(((((..(((((.......)))))..(((((....))..)))..............-.---)))))....(-(((......))))..... ( -18.20) >DroPse_CAF1 2907 99 + 1 CCACUGUACCAUGGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCGUCUCCCUUCCAGGCCCCCCUCCUAAUUCUA--UUGGUUAAU .......((((.((((((((.......(((...)))...(((....)))..))))))))((((.......))))................--.)))).... ( -23.60) >DroEre_CAF1 1991 91 + 1 CCACUGUA---GAGGCAUGCUCAUUAUGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCCUCCCAAU-C---AACACCCACC-AACCCUC--CUUGUCAGU ..((((.(---(.(((((((......((((...))))..(((....)))..))))))).))......-.---.........(-((.....--.))).)))) ( -17.40) >DroYak_CAF1 2165 93 + 1 CCACUGUA---GAGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCCUCCCAAU-C---ACCUCCCACC-AACCCUCCCCUUGUCAAU .......(---(.(((((((.......(((...)))...(((....)))..))))))).))......-.---..........-.................. ( -15.70) >DroAna_CAF1 2189 85 + 1 CCACUGUC---CAGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCCUCCCA-U-C---GCCA--------CCCCUUCUUGGGUUAAU ........---..(((((((.......(((...)))...(((....)))..))))))).......-.-.---...(--------(((......)))).... ( -19.20) >DroPer_CAF1 2897 99 + 1 CCACUGUACCAUGGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCGUCUCUCUUCCAGGCCCCCCUCCUAAUUCUA--UUGGUUAAU .......((((.((((((((.......(((...)))...(((....)))..))))))))((((.......))))................--.)))).... ( -23.60) >consensus CCACUGUA___GAGGCAUGCUCAUUACGACUGAGUCAGCAUCUAACGAUUUGCAUGCCCCUCCCAAU_C___GCCACCCACC_AACCCUC__UUGGUCAAU .............(((((((.......(((...)))...(((....)))..)))))))........................................... (-12.98 = -14.98 + 2.00)



| Location | 8,090,975 – 8,091,068 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -20.07 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8090975 93 - 23771897 AUUGACAACAGGGGGGUU-GGUGGGAGGC---G-AUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUC---UACAGUGG .....((((......)))-)........(---.-((((..((((((((((...((((((...((((...))))..)))))))))))))))---).)))).) ( -29.20) >DroPse_CAF1 2907 99 - 1 AUUAACCAA--UAGAAUUAGGAGGGGGGCCUGGAAGGGAGACGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCCAUGGUACAGUGG ....((((.--.....(((((.......)))))...(....)((((((((...((((((...((((...))))..)))))))))))))).))))....... ( -29.80) >DroEre_CAF1 1991 91 - 1 ACUGACAAG--GAGGGUU-GGUGGGUGUU---G-AUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCAUAAUGAGCAUGCCUC---UACAGUGG ((..((...--....)).-.)).......---.-((((..((((((((((...((((((...((((...))))..)))))))))))))))---).)))).. ( -30.30) >DroYak_CAF1 2165 93 - 1 AUUGACAAGGGGAGGGUU-GGUGGGAGGU---G-AUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUC---UACAGUGG ((..((.........)).-.)).......---.-((((..((((((((((...((((((...((((...))))..)))))))))))))))---).)))).. ( -27.30) >DroAna_CAF1 2189 85 - 1 AUUAACCCAAGAAGGGG--------UGGC---G-A-UGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUG---GACAGUGG ....((((......)))--------)..(---(-.-((....((((((((...((((((...((((...))))..)))))))))))))).---..)).)). ( -24.80) >DroPer_CAF1 2897 99 - 1 AUUAACCAA--UAGAAUUAGGAGGGGGGCCUGGAAGAGAGACGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCCAUGGUACAGUGG ....((((.--.....(((((.......))))).........((((((((...((((((...((((...))))..)))))))))))))).))))....... ( -28.70) >consensus AUUAACAAA__GAGGGUU_GGUGGGAGGC___G_AUUGGGAGGGGCAUGCAAAUCGUUAGAUGCUGACUCAGUCGUAAUGAGCAUGCCUC___UACAGUGG ..........................................((((((((...((((((...((((...))))..))))))))))))))............ (-20.07 = -19.85 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:03 2006