
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,049,673 – 8,049,897 |
| Length | 224 |
| Max. P | 0.978883 |

| Location | 8,049,673 – 8,049,793 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8049673 120 - 23771897 CAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAACACGUUUUUCUCCUUACCCCUGGCUGCACUCGACUUUUGUA .((((((((.......))))))))....(((((((...)))))))..................(((((((((((.....))))))))...((........))..)))............. ( -24.60) >DroSec_CAF1 20689 120 - 1 CAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAACACGUUUUUCACCUUACCCUUGGCUGCACUCGACUUUUGUG .((((((((.......)))))))).......((((((.(((((...((((.(((......))).))))((((((.....))))))............))))).)))...)))........ ( -25.20) >DroSim_CAF1 20846 120 - 1 CAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAACACGUUUUUCUCCUUACCCUUGGCUGCACUCGACUUUUGUG .((((((((.......)))))))).......((((((.(((((...((((.(((......))).))))((((((.....))))))............))))).)))...)))........ ( -25.20) >DroEre_CAF1 20620 120 - 1 CAUGCGUAAUUGAUUUUUAUGCAUUUUCCUAUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAACACGUUUUUCUCCUUACCCUUGGUUGCACUCGACUUUUGUG .((((((((.......)))))))).......((((((.(((((...((((.(((......))).))))((((((.....))))))............))))).)))...)))........ ( -25.40) >DroYak_CAF1 21272 120 - 1 CAUGCGUAAUUGAUUUUUAUGCAUUUCCCCUGCGGCAUCCGAGAGAUUGUUUAUUUAUUUGUAUGCAAAGCGUGCUUAACACGUUUUUCUCCUUACCCUUGGUUGCACUCGACUUGUGUG .((((((((.......))))))))...((..(.((.....((((((((((.(((......))).))))((((((.....))))))))))))....)))..))..((((.......)))). ( -25.80) >consensus CAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAACACGUUUUUCUCCUUACCCUUGGCUGCACUCGACUUUUGUG .((((((((.......))))))))..........(((.(((((...((((.(((......))).))))((((((.....))))))............))))).))).............. (-24.48 = -24.52 + 0.04)


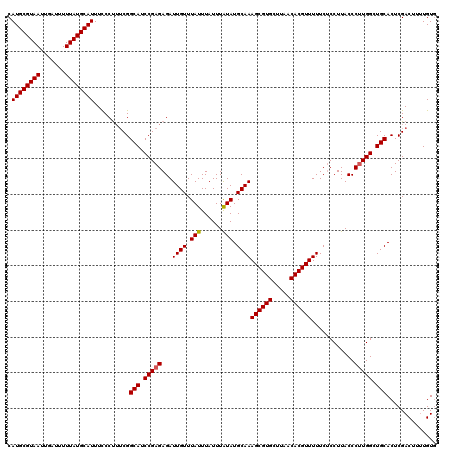
| Location | 8,049,713 – 8,049,828 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8049713 115 - 23771897 --UGGCUUAUCGUUA---AGUUUUUCUCAAUUGGAUUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAAC --.........((((---(((...........(((......((((((((.......)))))))).)))(((((((...))))))).((((.(((......))).)))).....))))))) ( -22.90) >DroSec_CAF1 20729 118 - 1 --UGGCUUAUCGUUAUUUUAUUUUUCUCAAUUGGAAUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAAC --.(((....((((..................((((.....((((((((.......))))))))))))(((((((...))))))).((((.(((......))).)))))))).))).... ( -23.60) >DroSim_CAF1 20886 115 - 1 --UGGCUUAUCGUUG---AAUUUUUCUCAAUUGGAAUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAAC --.(((....(((((---((..((((......))))..)))((((((((.......))))))))....(((((((...))))))).((((.(((......))).)))))))).))).... ( -25.40) >DroEre_CAF1 20660 114 - 1 ---GGCUUAUCGUUG---AAUUUUUCUCAAUUGGAAUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUUCCUAUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAAC ---(((....((((.---......((((....((((.....((((((((.......)))))))).))))..((((...))))))))((((.(((......))).)))))))).))).... ( -24.20) >DroYak_CAF1 21312 117 - 1 UUUGGCUUAUCGUUG---AGUUUUUCUCAAUUGGAAUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUCCCCUGCGGCAUCCGAGAGAUUGUUUAUUUAUUUGUAUGCAAAGCGUGCUUAAC ...(((.....((((---((.....)))))).((((.....((((((((.......))))))))))))...((.((((.((((((((.....)))).)))).))))...))..))).... ( -24.50) >consensus __UGGCUUAUCGUUG___AAUUUUUCUCAAUUGGAAUUUUCAUGCGUAAUUGAUUUUUAUGCAUUUCCCUUUCGGCAUCCGAGAGAUUGUUUAUUUAUUUAUAUGCAAAGCGUGCUUAAC ...(((....((((..........((((....((((.....((((((((.......))))))))))))...((((...))))))))((((.(((......))).)))))))).))).... (-22.12 = -22.20 + 0.08)

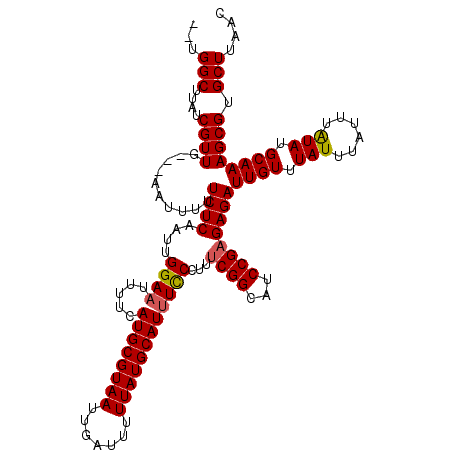

| Location | 8,049,793 – 8,049,897 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -6.28 |
| Energy contribution | -6.69 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8049793 104 + 23771897 AAAAAUCCAAUUGAGAAAAACU---UAACGAUAAGCCA---------UGGCGCAUGCGUGGCCUGAAGCUGAAGUCGGAAAAAAAAGUCG-GGCGUGGUAUGU---GUCUUGGUAAAAGU ....(((...(((((.....))---))).)))..((((---------.(((((((((...((((((..(((....))).........)))-)))...))))))---))).))))...... ( -29.80) >DroSec_CAF1 20809 104 + 1 AAAAUUCCAAUUGAGAAAAAUAAAAUAACGAUAAGCCA---------UGGCGCAUGCGUGGCCUGAAGGUGAAGUCCGAAAAAA---UUG-GGCGUGGUCUGU---GUCUUGGUAAAAGU ..................................((((---------.((((((.((.(.(((....))).).((((((.....---)))-)))...)).)))---))).))))...... ( -24.60) >DroSim_CAF1 20966 101 + 1 AAAAUUCCAAUUGAGAAAAAUU---CAACGAUAAGCCA---------UGGCGCAUGCGUGGCCUGAAGCUGAAGUCGGAAAAAA---UUG-GGCGUGGUCUGC---GUCUUGGUAAAAGU ..........(((((.....))---)))......((((---------.((((((.((...(((..(..(((....)))......---)..-)))...)).)))---))).))))...... ( -24.80) >DroEre_CAF1 20740 87 + 1 AAAAUUCCAAUUGAGAAAAAUU---CAACGAUAAGCC--------------------------CGAAGCUGAAGUCUUGAAGAA---UCG-GGCGUGGUAUCUCACAUUUUGGGAAAAGG ....(((((((((((.....))---))).((((.(((--------------------------(((..((..........))..---)))-)))....)))).......))))))..... ( -21.50) >DroYak_CAF1 21392 113 + 1 AAAAUUCCAAUUGAGAAAAACU---CAACGAUAAGCCAAACAAAUCAAGGCGCAUGCGUGGCCUCAAGCUGAAGUCUUGAAAAC---UUGGGGCGUGGUAUCUCAU-UUUUGGGAAAAGG ....((((((.(((((...((.---..(((....(((...........))).....))).((((((((...............)---)))))))))....))))).-..))))))..... ( -29.36) >consensus AAAAUUCCAAUUGAGAAAAAUU___CAACGAUAAGCCA_________UGGCGCAUGCGUGGCCUGAAGCUGAAGUCGGAAAAAA___UUG_GGCGUGGUAUGU___GUCUUGGUAAAAGU .........((..(((............((....(((...........(((.((....))))).......(....)...............))).))..........)))..))...... ( -6.28 = -6.69 + 0.41)

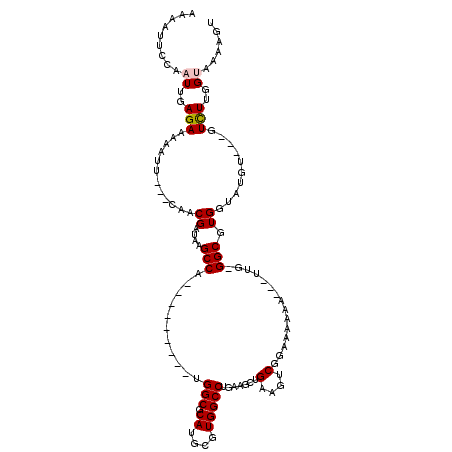
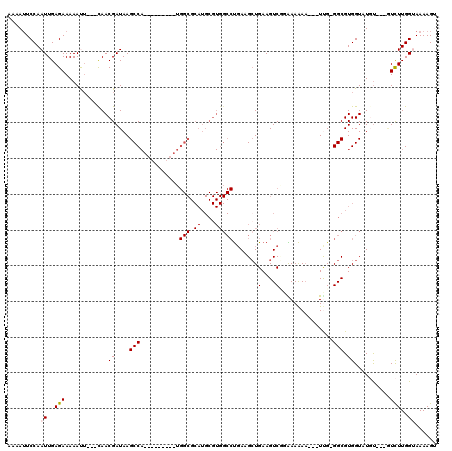
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:53 2006