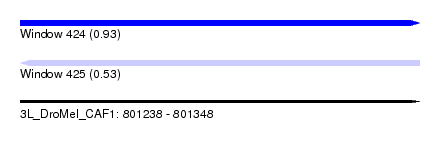
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 801,238 – 801,348 |
| Length | 110 |
| Max. P | 0.930150 |

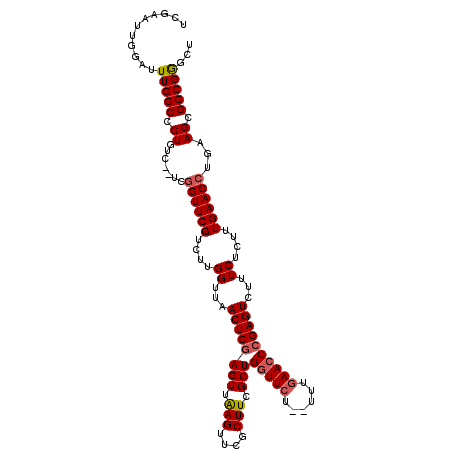
| Location | 801,238 – 801,348 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.87 |
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.71 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
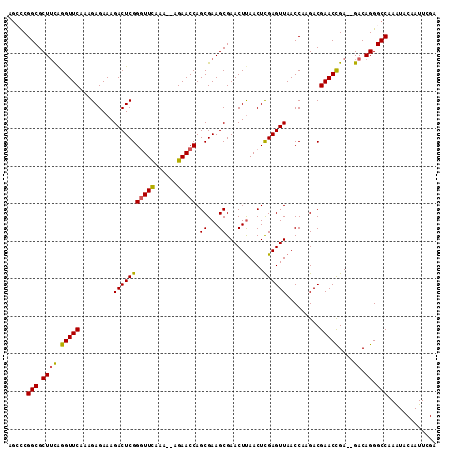
>3L_DroMel_CAF1 801238 110 + 23771897 AACCCGGCGCUUCAGGUUCAAAGAGAAAGACUCGGGUUCAAA--AGAACCAGCGAAGCGAACUUAACUCGAGUUAACCAAGACGAACCGA--GACAGGGCCAAAUACAAUUCGA .....(((.((((.(((((.........(((((((((((...--.)))))((..(((....)))..)))))))).........)))))..--)).)).)))............. ( -29.27) >DroSec_CAF1 164785 111 + 1 AGCCCGGCGCUUCAGGUUCAAAGAGAAAGACUCGGGUUCAAA--AGAACCAGCGAAGCGAACUUAACUCGAGUUAACCAAGACGAACCGAG-GACAGGGCCAAAUACAAUUCGA .((((....((((.(((((.........(((((((((((...--.)))))((..(((....)))..)))))))).........))))))))-)...)))).............. ( -32.47) >DroSim_CAF1 159439 112 + 1 AGCCCGGCGCUUCAGGUUCAAAGAGAAAGACUCGGGUUCAAA--AGAACCAGCGAAGCGAACUCAACUCGAGUUAACCAAGACGAACCGAGAGACAGGGCCAAAUGCAAUUCGA .((((((((((((.........(((.....))).(((((...--.)))))...))))))(((((.....)))))..)).........(....)...)))).............. ( -32.20) >DroEre_CAF1 164376 109 + 1 AGGCCGGCGCU-CGGGUUCAAAGAGAAAGACUCGGGUUCAAA--AGAACCAGCGAAGCGAACUUAACUUGAGUUAACCAAGACGAACCGA--GACAGGGCCAAAUCCAAUUCGA .((((....((-(((((((...(((.....))).(((((...--.)))))........))))....((((.(....))))).....))))--)....))))............. ( -33.00) >DroYak_CAF1 164313 112 + 1 AGAUCGGCGCUUUGGGUUCAAAGAGAAAGACUCGGUUUUGAACCCGAACCAGCGAAGCGAACUUAACUCGAGUUAACCAAGACGAACUGA--GACAGGGCCAAGUCCAAUUCGC .......((((((((((((((((.((.....))..))))))))))))...))))..(((((.....(((.((((..........))))))--)...((((...))))..))))) ( -32.60) >consensus AGCCCGGCGCUUCAGGUUCAAAGAGAAAGACUCGGGUUCAAA__AGAACCAGCGAAGCGAACUUAACUCGAGUUAACCAAGACGAACCGA__GACAGGGCCAAAUACAAUUCGA .....(((.((((.(((((.........(((((((((((......))))).((...))..........)))))).........)))))....)).)).)))............. (-24.95 = -24.71 + -0.24)

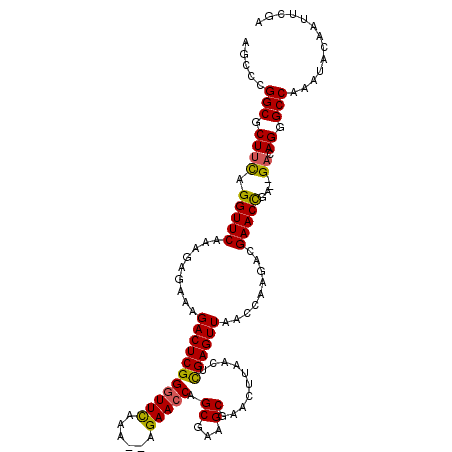
| Location | 801,238 – 801,348 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.87 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -25.06 |
| Energy contribution | -25.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 801238 110 - 23771897 UCGAAUUGUAUUUGGCCCUGUC--UCGGUUCGUCUUGGUUAACUCGAGUUAAGUUCGCUUCGCUGGUUCU--UUUGAACCCGAGUCUUUCUCUUUGAACCUGAAGCGCCGGGUU .........(((((((.((.((--..((((((....((...((((((((.(((....))).)))(((((.--...))))))))))...))....)))))).)))).))))))). ( -32.10) >DroSec_CAF1 164785 111 - 1 UCGAAUUGUAUUUGGCCCUGUC-CUCGGUUCGUCUUGGUUAACUCGAGUUAAGUUCGCUUCGCUGGUUCU--UUUGAACCCGAGUCUUUCUCUUUGAACCUGAAGCGCCGGGCU .............(((((.((.-(((((((((....((...((((((((.(((....))).)))(((((.--...))))))))))...))....)))))).).)).)).))))) ( -34.50) >DroSim_CAF1 159439 112 - 1 UCGAAUUGCAUUUGGCCCUGUCUCUCGGUUCGUCUUGGUUAACUCGAGUUGAGUUCGCUUCGCUGGUUCU--UUUGAACCCGAGUCUUUCUCUUUGAACCUGAAGCGCCGGGCU .............(((((((..((..((((((....((...((((((((.(((....))).)))(((((.--...))))))))))...))....)))))).))..))..))))) ( -34.60) >DroEre_CAF1 164376 109 - 1 UCGAAUUGGAUUUGGCCCUGUC--UCGGUUCGUCUUGGUUAACUCAAGUUAAGUUCGCUUCGCUGGUUCU--UUUGAACCCGAGUCUUUCUCUUUGAACCCG-AGCGCCGGCCU .............((((..(((--((((((((....((...((((.(((.(((....))).)))(((((.--...))))).))))...))....)))).)))-)).)).)))). ( -32.80) >DroYak_CAF1 164313 112 - 1 GCGAAUUGGACUUGGCCCUGUC--UCAGUUCGUCUUGGUUAACUCGAGUUAAGUUCGCUUCGCUGGUUCGGGUUCAAAACCGAGUCUUUCUCUUUGAACCCAAAGCGCCGAUCU (((((((((((........)))--.)))))))).(((((.....(.(((.(((....))).))).)...(((((((((...(((.....)))))))))))).....)))))... ( -35.30) >consensus UCGAAUUGGAUUUGGCCCUGUC__UCGGUUCGUCUUGGUUAACUCGAGUUAAGUUCGCUUCGCUGGUUCU__UUUGAACCCGAGUCUUUCUCUUUGAACCUGAAGCGCCGGGCU ...........(((((.((.......((((((....((...((((((((.(((....))).)))(((((......))))))))))...))....))))))...)).)))))... (-25.06 = -25.54 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:17 2006