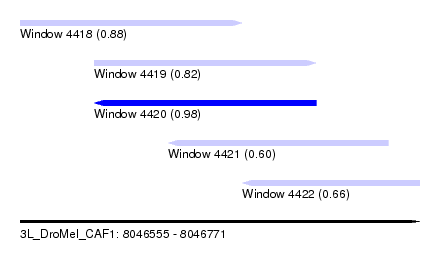
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,046,555 – 8,046,771 |
| Length | 216 |
| Max. P | 0.981404 |

| Location | 8,046,555 – 8,046,675 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046555 120 + 23771897 UUAUAAACAUUCUGAGGGAGCAAAGGUCAUGACUCACUUGGAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCC ............(((((((((((((((..(((.....(..(((.....)))..).(((((((...))))))).......))).))))))))..)))))))...((((....))))..... ( -28.20) >DroSec_CAF1 17678 118 + 1 UUAUAAACAUUCUCAGGGAGCAAAGGUCAUGACUCACUUGGAGCUGUUCUCUGGUUUUAUGG--ACAAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUAUCUUUAACAACCC ..............(((((((((((((..((((((.....))).(((((...........))--)))............))).))))))))..))))).....((((....))))..... ( -21.20) >DroSim_CAF1 17714 118 + 1 UUAUAAACAUUCUGAGGGAGCAAAGGUCAUGACUCACUUGGAGU--CUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCC ............(((((((((((((((...(((((.....))))--).....(..(((((((...)))))))..)........))))))))..)))))))...((((....))))..... ( -31.00) >DroEre_CAF1 17484 120 + 1 UUAUAAACAUUCAGUGGGAGCAAAGGUCAUGACUCACUUGGAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUUACAACCC .............(.((((((((((((..(((.....(..(((.....)))..).(((((((...))))))).......))).))))))))..)))).)..................... ( -22.60) >DroYak_CAF1 18046 120 + 1 UUAUAAACACACAGUGGGAGCAAAGGUCAUGACUCACUUGGAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUUACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUUACAACCC .............(.((((((((((((..(((.....(..(((.....)))..).(((((((...))))))).......))).))))))))..)))).)..................... ( -22.60) >consensus UUAUAAACAUUCUGAGGGAGCAAAGGUCAUGACUCACUUGGAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCC .............((((((((((((((..(((.....(..(((.....)))..).(((((((...))))))).......))).))))))))..))))))..................... (-22.28 = -23.08 + 0.80)

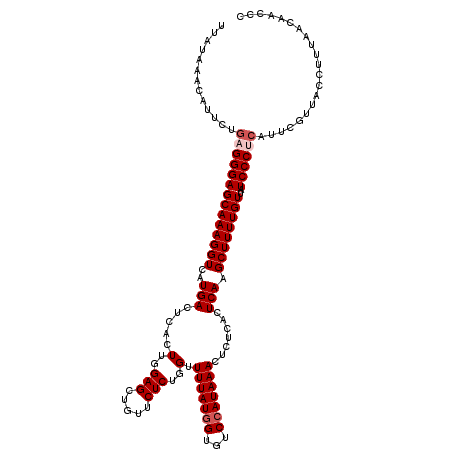
| Location | 8,046,595 – 8,046,715 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046595 120 + 23771897 GAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCCCUGAGCCACUUUUUAGGCUCAUUUUCAUUAUAGUUUUGCA (((((.......(..(((((((...)))))))..).......)))))((((((..................))))))....((((((........))))))................... ( -21.51) >DroSec_CAF1 17718 118 + 1 GAGCUGUUCUCUGGUUUUAUGG--ACAAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUAUCUUUAACAACCCCUGAGCCACUUUUUAGGCUCAUUUUGAUUAUAGUUUUGCA (((((((..((.((((....((--(.((((((..((......))..)))))).))).......((((....))))))))..((((((........))))))....))..))))))).... ( -24.90) >DroSim_CAF1 17754 118 + 1 GAGU--CUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCCCUGAGCCAGUUUUUAGGCUCAUUUUGAUUAUAGUUUUGCA ((((--......(..(((((((...)))))))..)..))))..((..((((.(((........((((....))))......((((((........))))))....))).))))....)). ( -22.20) >DroEre_CAF1 17524 120 + 1 GAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUUACAACCCCUGAGCCGCUUUUUAGGCUCAUUUUGAUUAUUGUUUUGCA (((((.......(..(((((((...)))))))..).......)))))..................................((((((........))))))................... ( -20.64) >DroYak_CAF1 18086 120 + 1 GAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUUACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUUACAACCCCUGAGCCGCUUUUUAGGCUCAUUUUGAUUAUUGUUUUGCA (((((.......(..(((((((...)))))))..).......)))))..................................((((((........))))))................... ( -20.64) >consensus GAGCUGUUCUCUGGUUUUAUGGUGUCCAUAAACUCUCACUCAAGCUUUUGUUAUCCCUCAUUCGUUACCUUUAACAACCCCUGAGCCACUUUUUAGGCUCAUUUUGAUUAUAGUUUUGCA (((((.......(..(((((((...)))))))..).......)))))((((((..................))))))....((((((........))))))................... (-18.33 = -18.97 + 0.64)


| Location | 8,046,595 – 8,046,715 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046595 120 - 23771897 UGCAAAACUAUAAUGAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUGAGUGAGAGUUUAUGGACACCAUAAAACCAGAGAACAGCUC ...................((((((........)))))).((((((((...(((..(......)((....((.((((((......)))))).))....)).....)))....)))))))) ( -26.90) >DroSec_CAF1 17718 118 - 1 UGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGAUAACGAAUGAGGGAUAACAAAAGCUUGAGUGAGAGUUUAUUGU--CCAUAAAACCAGAGAACAGCUC .((................((((((........))))))..((((((((....)))).......((((((.(((..(((....)))..))).))))--))....))))........)).. ( -26.80) >DroSim_CAF1 17754 118 - 1 UGCAAAACUAUAAUCAAAAUGAGCCUAAAAACUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUGAGUGAGAGUUUAUGGACACCAUAAAACCAGAGAG--ACUC (((......((((((....((((((........))))))..)))))).....)))........((.....((.((((((......)))))).))....))..............--.... ( -23.90) >DroEre_CAF1 17524 120 - 1 UGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAGGGGUUGUAAAAGGUAACGAAUGAGGGAUAACAAAAGCUUGAGUGAGAGUUUAUGGACACCAUAAAACCAGAGAACAGCUC .((......((((((....((((((........))))))..))))))....(((..(......)((....((.((((((......)))))).))....)).....)))........)).. ( -25.10) >DroYak_CAF1 18086 120 - 1 UGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAGGGGUUGUAAAAGGUAACGAAUGAGGGAUAACAAAAGCUUGAGUAAGAGUUUAUGGACACCAUAAAACCAGAGAACAGCUC .((......((((((....((((((........))))))..))))))....(((..(......)((....((.((((((......)))))).))....)).....)))........)).. ( -25.10) >consensus UGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUGAGUGAGAGUUUAUGGACACCAUAAAACCAGAGAACAGCUC ...................((((((........)))))).((((((((...(((..(......)((....((.((((((......)))))).))....)).....)))....)))))))) (-22.10 = -22.98 + 0.88)

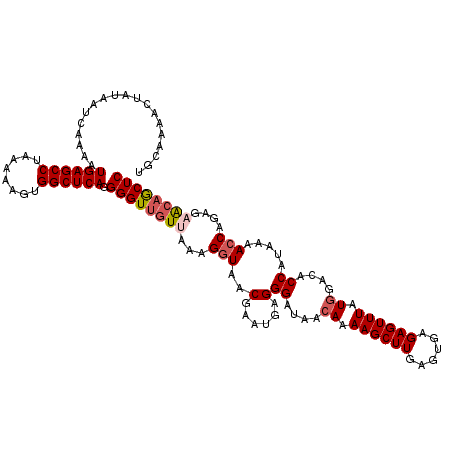
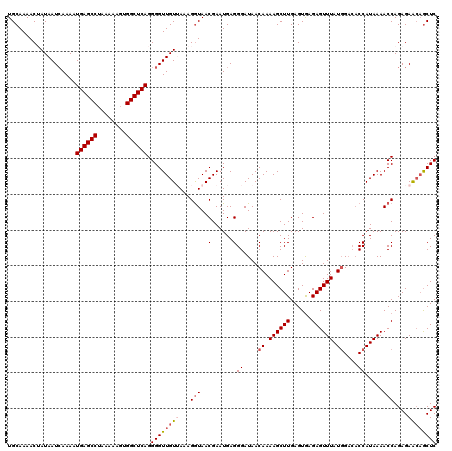
| Location | 8,046,635 – 8,046,754 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046635 119 - 23771897 AUUUUCUUAGUGCAGAACGCCAC-UUUUUCUUUAAGUGUAUGCAAAACUAUAAUGAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUG ..(..((((.((((......(((-((.......)))))..))))...((.(((..(...((((((........))))))....)..))).))........))))..)............. ( -25.30) >DroSec_CAF1 17756 119 - 1 AUAUCCCAAGUGCAGAACACCAU-UUUUUUUCUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGAUAACGAAUGAGGGAUAACAAAAGCUUG .((((((...((((..((((...-...........)))).)))).....((((((....((((((........))))))..))))))................))))))........... ( -29.74) >DroSim_CAF1 17792 120 - 1 AUUUCCCAAGUGCAGAACACCACUUUUUUUUCUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAACUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUG ...((((...((((......(((((........)))))..))))..(((((((((....((((((........))))))..))))))....))).........))))............. ( -27.90) >DroEre_CAF1 17564 119 - 1 AUUUCUCAGGCACAAGGCAGCAC-UUUUGUUUCAAGUGUAUGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAGGGGUUGUAAAAGGUAACGAAUGAGGGAUAACAAAAGCUUG ((..((((..(.....(((((((-((.......)))))).)))......((((((....((((((........))))))..))))))..........)..))))..))............ ( -31.60) >DroYak_CAF1 18126 118 - 1 AUUUCUCAGGUAGAAGACAGCAC-UUUUCUUU-AAGUGUAUGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAGGGGUUGUAAAAGGUAACGAAUGAGGGAUAACAAAAGCUUG ((..((((...........((((-((......-)))))).(((......((((((....((((((........))))))..)))))).....))).....))))..))............ ( -28.30) >consensus AUUUCCCAAGUGCAGAACACCAC_UUUUUUUUUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAGGGGUUGUUAAAGGUAACGAAUGAGGGAUAACAAAAGCUUG ...((((..((.....))..................(((.(((......((((((....((((((........))))))..)))))).....)))))).....))))............. (-19.56 = -19.08 + -0.48)

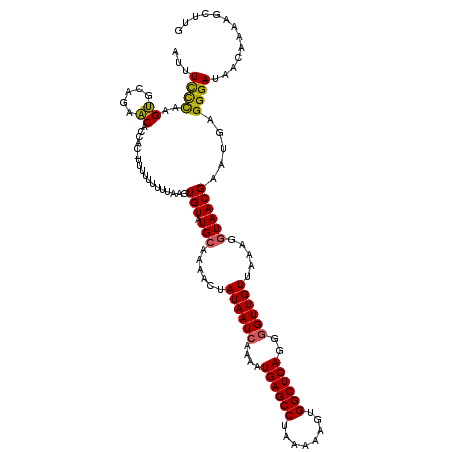
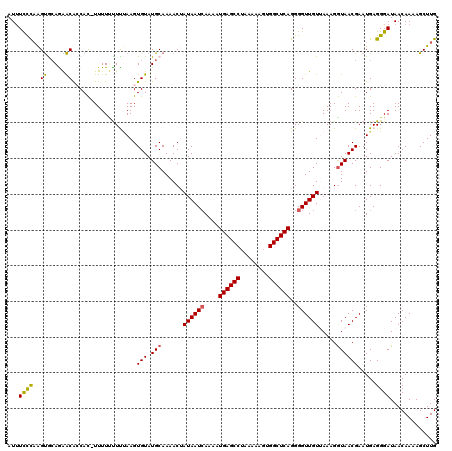
| Location | 8,046,675 – 8,046,771 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046675 96 - 23771897 ----AUUU----CUAGCUACUU-------U--------GUAUUUUCUUAGUGCAGAACGCCAC-UUUUUCUUUAAGUGUAUGCAAAACUAUAAUGAAAAUGAGCCUAAAAAGUGGCUCAG ----.(((----(.......((-------(--------(((((.....))))))))..(((((-((.......)))))...))...........)))).((((((........)))))). ( -21.20) >DroSec_CAF1 17796 104 - 1 ----AUUU----CUAAUUACUU-------UUCAGUACUAUAUAUCCCAAGUGCAGAACACCAU-UUUUUUUCUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAG ----....----.....((((.-------...))))..............((((..((((...-...........)))).))))...............((((((........)))))). ( -16.84) >DroSim_CAF1 17832 105 - 1 ----AUUU----CUAAUUACCU-------UUCAGUACUAUAUUUCCCAAGUGCAGAACACCACUUUUUUUUCUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAACUGGCUCAG ----....----..........-------(((.(((((..........))))).)))...(((((........))))).....................((((((........)))))). ( -17.90) >DroEre_CAF1 17604 119 - 1 GCACAUUUACGUCAAAUUACUGCUAAUACUGCAGUAUUGUAUUUCUCAGGCACAAGGCAGCAC-UUUUGUUUCAAGUGUAUGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAG ((..........(((..((((((.......)))))))))..........)).....(((((((-((.......)))))).)))................((((((........)))))). ( -27.75) >DroYak_CAF1 18166 107 - 1 ----AAUUACGGCUUAUUACUG-------UGCACUAUUAUAUUUCUCAGGUAGAAGACAGCAC-UUUUCUUU-AAGUGUAUGCAAAACAAUAAUCAAAAUGAGCCUAAAAAGCGGCUCAG ----......((.(((((....-------((((........(((((.....)))))...((((-((......-)))))).))))....)))))))....((((((........)))))). ( -22.40) >consensus ____AUUU____CUAAUUACUU_______UGCAGUACUAUAUUUCCCAAGUGCAGAACACCAC_UUUUUUUUUAAGUGUAUGCAAAACUAUAAUCAAAAUGAGCCUAAAAAGUGGCUCAG ..................................................((((..((((...............)))).))))...............((((((........)))))). (-10.98 = -11.86 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:48 2006