| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,046,300 – 8,046,419 |
| Length | 119 |
| Max. P | 0.693115 |

| Location | 8,046,300 – 8,046,419 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
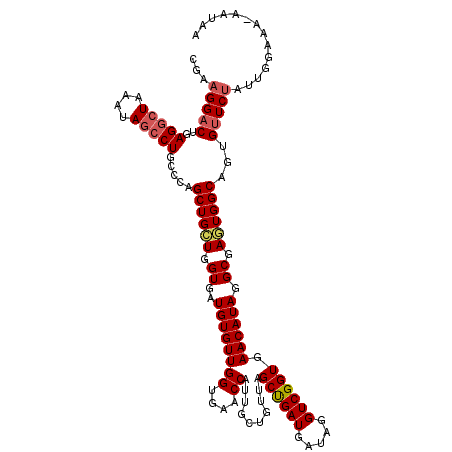
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -22.06 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046300 119 + 23771897 UUAUU-UUUCCCAUAGAACAUUGCCACUCGCCUAUGUUUACCGACCUAUCAUCAGCUAAUAGCAAUGGUUCACCAACACAUCACCAGCAGCUGGGCAGGCUAUUCAGCCUCAGUCCUUCG .....-.........(((((((((.....((..(((.............)))..)).....))))).))))..................(..(((((((((....)))))..))))..). ( -21.62) >DroSec_CAF1 17407 119 + 1 UUAUU-UUUCCAAUAGAACACUGCCACUCGCCUAUGUUCACCGACAUAUCAUCAGCUAACAGCAAUGGUUCACCAACACAUCACCAGCAGCUGGGCAGGCUAUUUAGCCUCAGUCCGUCG .....-.........(((((..((.....))...)))))..((((...............(((..((((.............))))...)))(((((((((....)))))..)))))))) ( -26.42) >DroSim_CAF1 17443 119 + 1 CUAUU-UUUCCAAUAGAACACUGCCACUCGCCUAUGUUCACCGACCUAUCAUCAGCUAACAGCAAUGGUUCACCAACACAUCACCAGCAGCUGGGCAGGCUAUUUAGUCUCAGUCCUUCC .....-.....(((((....(((((....((..(((.............)))..))...((((..((((.............))))...))))))))).)))))................ ( -21.84) >DroEre_CAF1 17205 119 + 1 UUCUU-UCUUCAAUAGAGCACUGCCAGUCGCCUAUGUUCACCGACUUAUCAUCAGCUAACAGCAAUGGUUCACCAACACAUCACCACCAGCUGGGCAGGUUGUUUAGCCUCAGUCCUUCG .....-.........(((((((((((((((...........))))).............((((..((((.............))))...)))))))))..)))))............... ( -22.12) >DroYak_CAF1 17777 120 + 1 UUGUUCUUUUUAAUAGAACACAGCCACUCGCCUAUGUUCACCGACAUAUCAUCGGCGAACAGCAAUGGUUCACCAACACAUCACCACCAGCUGGGCAGGCUAUUCAGCCUCAGUCCUUCG .((((((.......))))))..(((..(((((((((((....)))))).....))))).((((..((((.............))))...)))))))(((((....))))).......... ( -33.22) >consensus UUAUU_UUUCCAAUAGAACACUGCCACUCGCCUAUGUUCACCGACAUAUCAUCAGCUAACAGCAAUGGUUCACCAACACAUCACCAGCAGCUGGGCAGGCUAUUUAGCCUCAGUCCUUCG ...............(((((..((.....))...)))))...(((.........((...((((..((((.............))))...)))).))(((((....)))))..)))..... (-22.06 = -21.26 + -0.80)

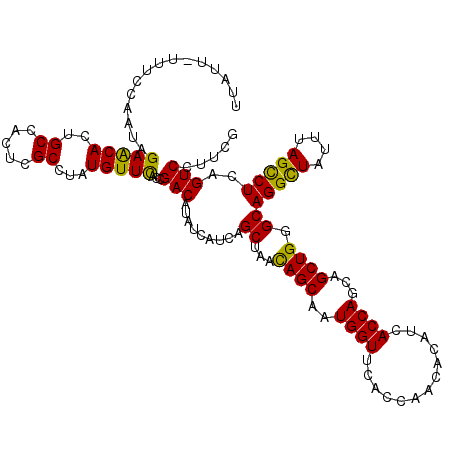
| Location | 8,046,300 – 8,046,419 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8046300 119 - 23771897 CGAAGGACUGAGGCUGAAUAGCCUGCCCAGCUGCUGGUGAUGUGUUGGUGAACCAUUGCUAUUAGCUGAUGAUAGGUCGGUAAACAUAGGCGAGUGGCAAUGUUCUAUGGGAAA-AAUAA ((.(((((..(((((....)))))(((((((((.(((..(((.(((....))))))..))).)))))).......(((.(....)...)))....)))...))))).)).....-..... ( -35.20) >DroSec_CAF1 17407 119 - 1 CGACGGACUGAGGCUAAAUAGCCUGCCCAGCUGCUGGUGAUGUGUUGGUGAACCAUUGCUGUUAGCUGAUGAUAUGUCGGUGAACAUAGGCGAGUGGCAGUGUUCUAUUGGAAA-AAUAA ((((((....(((((....))))).))((((((..((..(((.(((....))))))..))..)))))).......))))..((((((..((.....)).)))))).........-..... ( -37.00) >DroSim_CAF1 17443 119 - 1 GGAAGGACUGAGACUAAAUAGCCUGCCCAGCUGCUGGUGAUGUGUUGGUGAACCAUUGCUGUUAGCUGAUGAUAGGUCGGUGAACAUAGGCGAGUGGCAGUGUUCUAUUGGAAA-AAUAG ((((.(.(((..(((.....(((((..((((((..((..(((.(((....))))))..))..))))))...........(....).))))).)))..)))).))))........-..... ( -32.70) >DroEre_CAF1 17205 119 - 1 CGAAGGACUGAGGCUAAACAACCUGCCCAGCUGGUGGUGAUGUGUUGGUGAACCAUUGCUGUUAGCUGAUGAUAAGUCGGUGAACAUAGGCGACUGGCAGUGCUCUAUUGAAGA-AAGAA .((.(.(((.(((........)))((((((((((..(..(((.(((....))))))..)..)))))))......(((((.(.......).))))))))))).))).........-..... ( -35.50) >DroYak_CAF1 17777 120 - 1 CGAAGGACUGAGGCUGAAUAGCCUGCCCAGCUGGUGGUGAUGUGUUGGUGAACCAUUGCUGUUCGCCGAUGAUAUGUCGGUGAACAUAGGCGAGUGGCUGUGUUCUAUUAAAAAGAACAA ....((....(((((....))))).))((((((.(.((...((..(((....)))..))(((((((((((.....)))))))))))...)).).))))))((((((.......)))))). ( -43.20) >consensus CGAAGGACUGAGGCUAAAUAGCCUGCCCAGCUGCUGGUGAUGUGUUGGUGAACCAUUGCUGUUAGCUGAUGAUAGGUCGGUGAACAUAGGCGAGUGGCAGUGUUCUAUUGGAAA_AAUAA ...(((((..(((((....))))).....((((((.((..((((((((....))..........((((((.....)))))).)))))).)).))))))...))))).............. (-29.98 = -30.58 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:43 2006