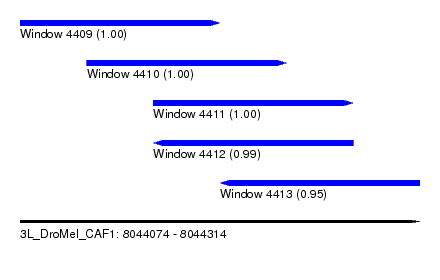
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,044,074 – 8,044,314 |
| Length | 240 |
| Max. P | 0.997774 |

| Location | 8,044,074 – 8,044,194 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -29.56 |
| Energy contribution | -32.20 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8044074 120 + 23771897 AGAUAUUGACACGCCCCGUCAAUAUAAAUAAUUGCUAAUUGCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUUGACUCGCUCAAUU ..((((((((.......))))))))........((....((((((.(((.....))).)))))).......(((((.((((...((((....))))...))))....)))))))...... ( -33.10) >DroSec_CAF1 15171 120 + 1 AGAUAUUGACACGCCCCGUCAAUAUAAAUAAUUGCUAAUUGCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUUCGGCUCCGAAAACAUUCGACUCGCUCAAUU ..((((((((.......))))))))........((....((((((.(((.....))).)))))).......(((((.((((...((((....))))...))))....)))))))...... ( -33.60) >DroSim_CAF1 15174 120 + 1 AGAUAUUGACACGCCCCGUCAAUAUAAAUAAUUGCUAAUUGCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUU ..((((((((.......))))))))........((....((((((.(((.....))).)))))).......(((((.((((...((((....))))...))))....)))))))...... ( -33.10) >DroEre_CAF1 15000 120 + 1 AGAUAUUGACACGCCCCAUCAAUAUAAAUAAUUGCUAAUUGCUGGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUU ..(((((((.........)))))))........((....(((..(.(((.....))).)..))).......(((((.((((...((((....))))...))))....)))))))...... ( -28.00) >DroYak_CAF1 15472 120 + 1 AGAUAUUGACACGCCCCGUCAAUAUAAAUAAUUGCUAAUUGCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUU ..((((((((.......))))))))........((....((((((.(((.....))).)))))).......(((((.((((...((((....))))...))))....)))))))...... ( -33.10) >consensus AGAUAUUGACACGCCCCGUCAAUAUAAAUAAUUGCUAAUUGCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUU ..((((((((.......))))))))........((....((((((.(((.....))).)))))).......(((((.((((...((((....))))...))))....)))))))...... (-29.56 = -32.20 + 2.64)


| Location | 8,044,114 – 8,044,234 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -31.52 |
| Energy contribution | -34.52 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8044114 120 + 23771897 GCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUUGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...(((((((((.((((((....)))))).....)))))))))... ( -35.40) >DroSec_CAF1 15211 120 + 1 GCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUUCGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...(((((((((.((((((....)))))).....)))))))))... ( -35.90) >DroSim_CAF1 15214 120 + 1 GCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCACUUUAUUGUAAAUCCGUGUGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...(((((((((.(((((......))))).....)))))))))... ( -34.60) >DroEre_CAF1 15040 120 + 1 GCUGGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGCGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...((((((.((.((((((....)))))).....)).))))))... ( -30.60) >DroYak_CAF1 15512 120 + 1 GCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...(((((((((.((((((....)))))).....)))))))))... ( -35.40) >consensus GCUAGCCAUGAUUAAUGUCUGGCAUCUUUUGGAGUUCGUUUGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUG ((..((((.(((....)))))))........(((((.((((...((((....))))...))))....)))))))...(((((((((.((((((....)))))).....)))))))))... (-31.52 = -34.52 + 3.00)



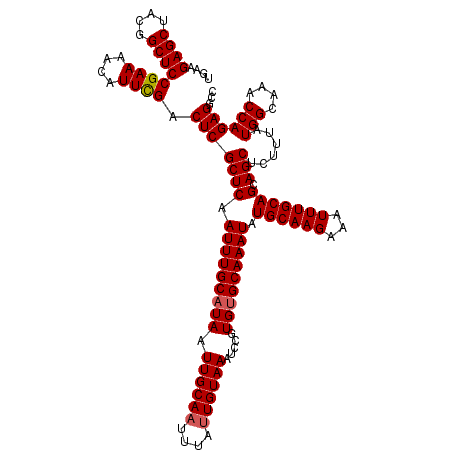
| Location | 8,044,154 – 8,044,274 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8044154 120 + 23771897 UGAAGAGCUACGGCUCCGAAAACAUUUGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC (((((((((.((....))..............((...(((((((((.((((((....)))))).....))))))))).((((((...))))))))))))))))).(((.........))) ( -31.20) >DroSec_CAF1 15251 120 + 1 UGAAGAGCUUCGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC ....((((....))))((((....)))).(((((((.(((((((((.((((((....)))))).....))))))))).((((((...))))))).)))......(((....))))))... ( -32.60) >DroSim_CAF1 15254 120 + 1 UGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCACUUUAUUGUAAAUCCGUGUGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC ....((((....))))((((....)))).(((((((((((((((((.((((((................)))))).))))))))....))).))).(((......)))......)))... ( -34.99) >DroEre_CAF1 15080 120 + 1 UGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGCGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC ....((((....))))((((....)))).(((((((((((((((((.((((....................)))).))))))))....))).))).(((......)))......)))... ( -29.95) >DroYak_CAF1 15552 120 + 1 UGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC ....((((....))))((((....)))).(((((((.(((((((((.((((((....)))))).....))))))))).((((((...))))))).)))......(((....))))))... ( -32.10) >consensus UGAAGAGCUACGGCUCCGAAAACAUUCGACUCGCUCAAUUUGCAUAAUUGCAAUUUAUUGUAAAUCCGUGUGCAAAUAUGCAAGAAAUUUGCAGCAGCUCUUUAUGGCAAACCAGAGGCC (((((((((.(.((..((((....)))).((.((...(((((((((.((((((....)))))).....)))))))))..)).))......)).).))))))))).(((.........))) (-30.20 = -30.44 + 0.24)


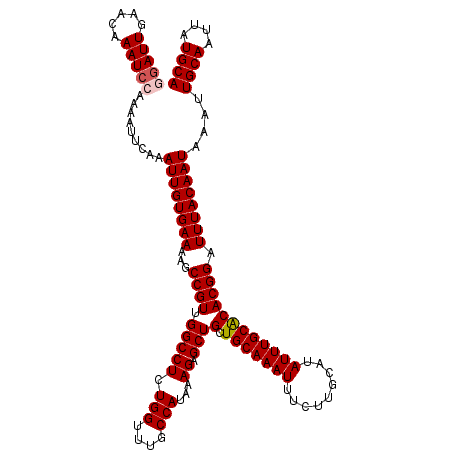
| Location | 8,044,154 – 8,044,274 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -32.59 |
| Energy contribution | -32.83 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8044154 120 - 23771897 GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCAAAUUGAGCGAGUCAAAUGUUUUCGGAGCCGUAGCUCUUCA (((.........)))...(((((((((.((.......(((((((.(((((..................))))).)))))))......((((.........))))..)).))))))))).. ( -32.77) >DroSec_CAF1 15251 120 - 1 GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCAAAUUGAGCGAGUCGAAUGUUUUCGGAGCCGAAGCUCUUCA (((.........)))...(((((((............(((((((.(((((..................))))).))))))).(((.((...(((((....))))).)))))))))))).. ( -30.97) >DroSim_CAF1 15254 120 - 1 GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAGUGCAAUUAUGCAAAUUGAGCGAGUCGAAUGUUUUCGGAGCCGUAGCUCUUCA (((.........)))...(((((((((.((.......(((((((.((((((................)))))).)))))))..........(((((....))))).)).))))))))).. ( -37.99) >DroEre_CAF1 15080 120 - 1 GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCGCACGGAUUUACAAUAAAUUGCAAUUAUGCAAAUUGAGCGAGUCGAAUGUUUUCGGAGCCGUAGCUCUUCA (((.........)))...(((((((((.((.......(((((((.((((.....((((((...)))))))))).)))))))..........(((((....))))).)).))))))))).. ( -34.00) >DroYak_CAF1 15552 120 - 1 GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCAAAUUGAGCGAGUCGAAUGUUUUCGGAGCCGUAGCUCUUCA (((.........)))...(((((((((.((.......(((((((.(((((..................))))).)))))))..........(((((....))))).)).))))))))).. ( -34.37) >consensus GGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCAAAUUGAGCGAGUCGAAUGUUUUCGGAGCCGUAGCUCUUCA (((.........)))...(((((((((.((.......(((((((.(((((..................))))).)))))))..........(((((....))))).)).))))))))).. (-32.59 = -32.83 + 0.24)



| Location | 8,044,194 – 8,044,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8044194 120 - 23771897 GGAUUGAACAAAUCCAAUAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCA (((((.....))))).........((((((((...((((.(((((.(((....)))...)).)))..(((((((.........))))))).)))).))))))))....((((....)))) ( -28.50) >DroSec_CAF1 15291 120 - 1 GGAUUGAACAAAUCCAAAAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCA (((((.....))))).........((((((((...((((.(((((.(((....)))...)).)))..(((((((.........))))))).)))).))))))))....((((....)))) ( -28.50) >DroSim_CAF1 15294 120 - 1 GGAUUGAACAAAUCCAAAAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAGUGCAAUUAUGCA (((((.....)))))..........(((((..((((((........))))))..)))))...((....))........((((((.((((((................)))))).)))))) ( -29.19) >DroEre_CAF1 15120 120 - 1 AGAUUGAAAAAAUCCAAAAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCGCACGGAUUUACAAUAAAUUGCAAUUAUGCA .((((.....))))..........((((((((...((((.(((((.(((....)))...)).)))((.((((((.........)))))))))))).))))))))....((((....)))) ( -26.00) >DroYak_CAF1 15592 120 - 1 AGAUUGAAAAAAUACAAAAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCA ...(((((...........)))))((((((((...((((.(((((.(((....)))...)).)))..(((((((.........))))))).)))).))))))))....((((....)))) ( -23.90) >consensus GGAUUGAACAAAUCCAAAAUUCAAAUUGUGAAAAGCCGUUGGCCUCUGGUUUGCCAUAAAGAGCUGCUGCAAAUUUCUUGCAUAUUUGCACACGGAUUUACAAUAAAUUGCAAUUAUGCA (((((.....))))).........((((((((...((((.(((((.(((....)))...)).)))(.(((((((.........)))))))))))).))))))))....((((....)))) (-25.30 = -25.74 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:39 2006