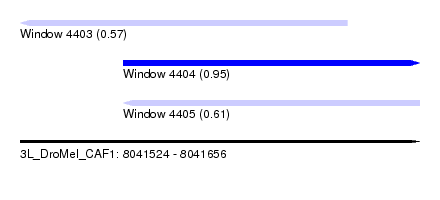
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,041,524 – 8,041,656 |
| Length | 132 |
| Max. P | 0.949402 |

| Location | 8,041,524 – 8,041,632 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
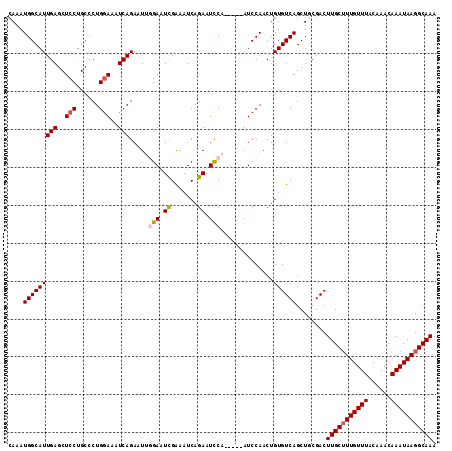
>3L_DroMel_CAF1 8041524 108 - 23771897 CAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUUGGAAUCAGAAUCCA-----AUCCAACUGUGUCAGCUGCGACUUGCUUUGUUUACAAACAAAUAAGGCAAA ((..(((((((((..(((......)))..)))...(((((.(((((.......))))-----))))))..))))))..))....(((((((((((......))))))))))). ( -30.70) >DroSec_CAF1 12653 108 - 1 CAAAUGGCAUUGAGCUCCUGCCCUGCAAAUCAGAAUUGGAAUCGAAAUCAGAAUCCA-----AUCCAACUGUGUCAGCUGCGACUUGCUUUGUUUACAAACAAAUAAGGCAAA ((..((((((.(.((.........))......(.((((((.((.......)).))))-----)).)..).))))))..))....(((((((((((......))))))))))). ( -24.60) >DroSim_CAF1 12669 108 - 1 CAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUCGAAAUCAGAAUUCA-----AUCCAACUGUGUCAGCUGCGACUUGCUUUGUUUACAAACAAAUAAGGCAAA ((..(((((((((..(((......)))..)))(.((((((.((.......)).))))-----)).)....))))))..))....(((((((((((......))))))))))). ( -26.00) >DroEre_CAF1 12490 108 - 1 CAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUCAGAGUCGAAAUCAGAAUCCA-----AUCCAACUGUGUCAGCUUCGACUUGCGUUGUUUACAAACAAAUAAGGCAAA ....(((..((((..(((......)))..))))..)))((((((((....((...((-----.......))..))...))))))))((.((((((......)))))).))... ( -21.80) >DroYak_CAF1 12972 113 - 1 CAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGACUCAGAAUCGAAAUCAGAAUCCAAUCCAAUCCAACUGUGUCAGCUUCGACUUGCUUUGUUUACAAACAAAUAAGGCAAA .....(((..(((..(((......)))..)))(((.(((....(....).((......))........))).))).))).....(((((((((((......))))))))))). ( -22.80) >consensus CAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUCGAAAUCAGAAUCCA_____AUCCAACUGUGUCAGCUGCGACUUGCUUUGUUUACAAACAAAUAAGGCAAA ....(((((((((..(((......)))..))).....(((.((.......)).)))..............))))))........(((((((((((......))))))))))). (-19.76 = -20.24 + 0.48)

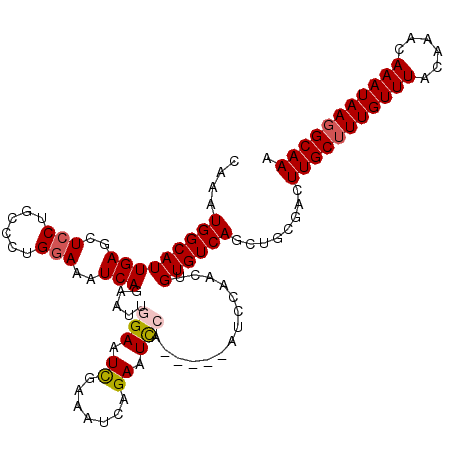
| Location | 8,041,558 – 8,041,656 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
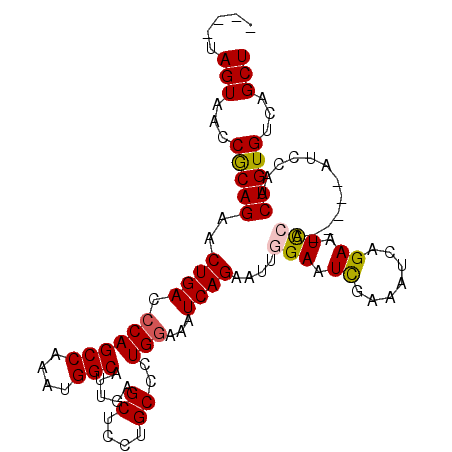
>3L_DroMel_CAF1 8041558 98 + 23771897 AGCUGACACAGUUGGAU-----UGGAUUCUGAUUCCAAUUCCAAUUCUGAUUUCCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGCGGUUACUA---- (((((((.(((((((((-----((((.......))))).)))))..)))....(((((((....))))...(((....)))))))))))))............---- ( -32.50) >DroSec_CAF1 12687 98 + 1 AGCUGACACAGUUGGAU-----UGGAUUCUGAUUUCGAUUCCAAUUCUGAUUUGCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGCGGUUACUA---- ((.((((......((((-----((((.((.......)).)))))))).....((((((((..((.((((..(((....))))))))).)))))))))))))).---- ( -31.30) >DroSim_CAF1 12703 98 + 1 AGCUGACACAGUUGGAU-----UGAAUUCUGAUUUCGAUUCCAAUUCUGAUUUCCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGCGGUUACUA---- (((((((.(((((((((-----((((.......))))).)))))..)))....(((((((....))))...(((....)))))))))))))............---- ( -29.80) >DroEre_CAF1 12524 98 + 1 AGCUGACACAGUUGGAU-----UGGAUUCUGAUUUCGACUCUGAUUCUGAUUUCCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGUGGUUACUA---- (((((((.((((..(((-----.((....(((((((..(((((...........)))))..)))).)))...)))))..)))).)))))))............---- ( -31.00) >DroYak_CAF1 13006 107 + 1 AGCUGACACAGUUGGAUUGGAUUGGAUUCUGAUUUCGAUUCUGAGUCUGAUUUCCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGUGGUUACUAUGUA (((((((.((((..((((((((..(((((.((.......)).)))))..)..)))))((((.........)))).))..)))).)))))))................ ( -35.70) >consensus AGCUGACACAGUUGGAU_____UGGAUUCUGAUUUCGAUUCCAAUUCUGAUUUCCAGGGCAGGAGCUCAAUGCCAUUUGGCUGGGUCAGUUCUGCGGUUACUA____ (((((((.((((..(((......(((.......)))((((((..(((((.....)))))..)))).))......)))..)))).)))))))................ (-26.22 = -25.90 + -0.32)

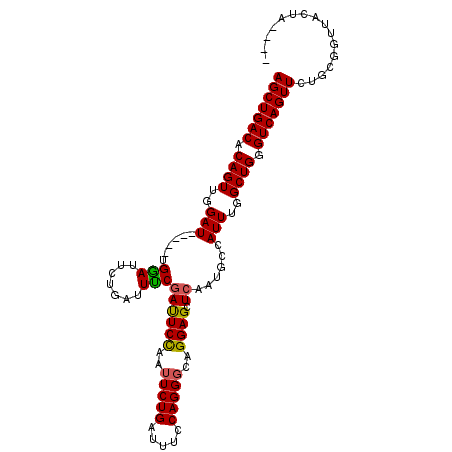
| Location | 8,041,558 – 8,041,656 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8041558 98 - 23771897 ----UAGUAACCGCAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUUGGAAUCAGAAUCCA-----AUCCAACUGUGUCAGCU ----.(((...(((((..((((.((((((....))).....((....))..)))...))))..(((((.(((((.......))))-----)))))))))))...))) ( -27.70) >DroSec_CAF1 12687 98 - 1 ----UAGUAACCGCAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGCAAAUCAGAAUUGGAAUCGAAAUCAGAAUCCA-----AUCCAACUGUGUCAGCU ----.(((...(((((..((((....(((....))).(((.((....))....))).)))).((((((.((.......)).))))-----))....)))))...))) ( -23.30) >DroSim_CAF1 12703 98 - 1 ----UAGUAACCGCAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUCGAAAUCAGAAUUCA-----AUCCAACUGUGUCAGCU ----..............(((((.(((((....)))..(((..(((......)))..)))(.((((((.((.......)).))))-----)).)...)).))))).. ( -22.10) >DroEre_CAF1 12524 98 - 1 ----UAGUAACCACAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUCAGAGUCGAAAUCAGAAUCCA-----AUCCAACUGUGUCAGCU ----.(((...(((((..((((....(((....))).((((..(((......)))..))))..))))..((.......)).....-----......)))))...))) ( -21.60) >DroYak_CAF1 13006 107 - 1 UACAUAGUAACCACAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGACUCAGAAUCGAAAUCAGAAUCCAAUCCAAUCCAACUGUGUCAGCU .....(((...(((((..((((....(((....))).((((..(((......)))..))))..))))..((.......))................)))))...))) ( -21.50) >consensus ____UAGUAACCGCAGAACUGACCCAGCCAAAUGGCAUUGAGCUCCUGCCCUGGAAAUCAGAAUUGGAAUCGAAAUCAGAAUCCA_____AUCCAACUGUGUCAGCU .....(((...(((((..((((.((((((....))).....((....))..)))...))))....(((.((.......)).)))............)))))...))) (-18.70 = -18.74 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:31 2006