
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,039,904 – 8,040,045 |
| Length | 141 |
| Max. P | 0.810225 |

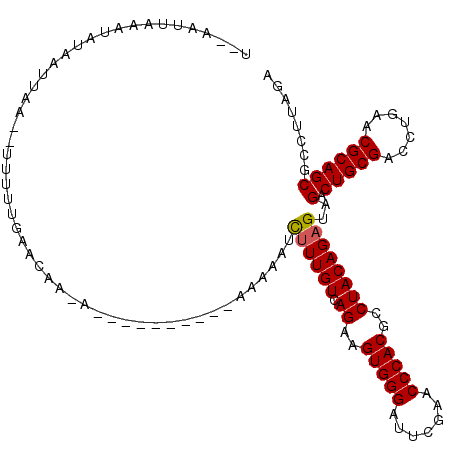
| Location | 8,039,904 – 8,040,005 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8039904 101 + 23771897 U--AAUUAAAUAUAAUACA--UUUUUGAACA-------------AAAAAUCUUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAGUAGACUGCGACCUGAACGCAGCGCCUUAGA .--................--.((((((.((-------------((.....))))))))))(((((.......))))).......(((..(.(((((.......))))))..)))... ( -21.50) >DroSec_CAF1 11072 104 + 1 U--AAUUAAAUGUAAUUAA--UUUUUGAACAACA----------AAAAAUCUUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAGUAGACUGCGACCUGAACGCAGCGCCUUAGA (--(((((....)))))).--((((((.....))----------))))..((((((.((..(((((.......)))))..))))))))..(.(((((.......))))))........ ( -23.00) >DroEre_CAF1 10880 118 + 1 UACAAUUAAAUCUAAUUAAUACUUUUGAACAAGAAAAGUUAAAAAAAAAUUCUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAGUAGACUGCGACCUGAACGCAGCGCCUUAGA ..........(((((.......((((((.((((((...((....))...))))))))))))(((((.......)))))((((((...)))).(((((.......))))).)).))))) ( -26.10) >consensus U__AAUUAAAUAUAAUUAA__UUUUUGAACAA_A__________AAAAAUCUUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAGUAGACUGCGACCUGAACGCAGCGCCUUAGA ..................................................((((((.((..(((((.......)))))..))))))))..(.(((((.......))))))........ (-19.32 = -19.43 + 0.11)


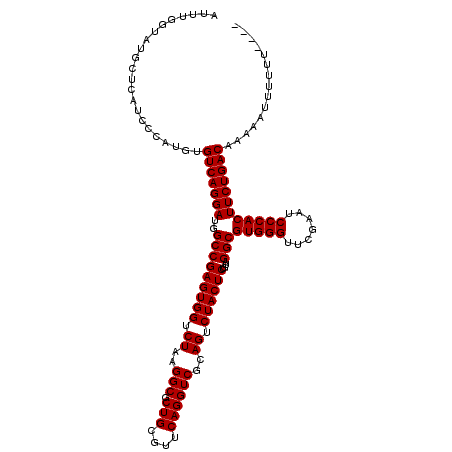
| Location | 8,039,931 – 8,040,045 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -35.97 |
| Energy contribution | -35.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8039931 114 - 23771897 AUUUGGUAUGCUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAGAUUUUU----- ...(((.((....)).)))..((((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))).........----- ( -38.10) >DroGri_CAF1 10236 116 - 1 GCUGGCGCACCUCAUGCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAGUUUUUUUG--- (.(((((.......))))).).(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))............--- ( -41.10) >DroSec_CAF1 11102 114 - 1 AUUUUGUAUACUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAGAUUUUU----- .....................((((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))).........----- ( -36.80) >DroEre_CAF1 10920 118 - 1 AUUUGGUAUGCUCAUCUUAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGAAUUUUUUUU-U .....................((((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......)))))))))))))............-. ( -36.80) >DroYak_CAF1 11376 119 - 1 AUUUGGUAUGCUCAUCUCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGAAUUUUUUGAAA ...........(((.......((((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))).........))).. ( -37.59) >DroMoj_CAF1 9693 116 - 1 UUUGGCUCAACUCAUGCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGAUUUUUUUU--- .(((((.........)))))..(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))............--- ( -41.60) >consensus AUUUGGUAUGCUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAAAUUUUUU____ ......................(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))............... (-35.97 = -35.97 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:27 2006