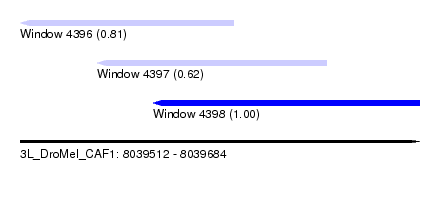
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,039,512 – 8,039,684 |
| Length | 172 |
| Max. P | 0.996713 |

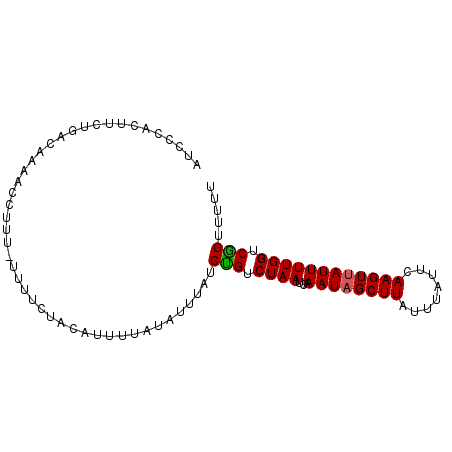
| Location | 8,039,512 – 8,039,604 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -9.41 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8039512 92 - 23771897 AUCCCACUUCUGACAAAACCUUU-UUUUAUCCCUUUUAUAUUUAUCUGUCUAAUCAAAUAGCUUAUUUAUUCAAGUUAUUUUGGUCAGUUUUU .......................-.....................(((.((((...((((((((........)))))))))))).)))..... ( -9.50) >DroEre_CAF1 10539 92 - 1 AUCCCACUUCUGACAAAACCUUU-UUUUCUACAUUUUGUAUUUAUCCGUCUAAUCAAAUAGCUUAUUUAUUCAAGUUAUUUUGGUCGGUUUUU ............((((((.....-.........))))))......(((.((((...((((((((........)))))))))))).)))..... ( -11.84) >DroYak_CAF1 10972 93 - 1 AUCCCACUUCUGACAAAUCCUUUUUUUUCUACUUUUUAUAUUUAUCUGUCUAAUCAAAUUGCUUAUUUAUUCAAGUUAUUUUGGUCGGUUUUU ...((......((((...............................))))..((((((..((((........))))...)))))).))..... ( -6.88) >consensus AUCCCACUUCUGACAAAACCUUU_UUUUCUACAUUUUAUAUUUAUCUGUCUAAUCAAAUAGCUUAUUUAUUCAAGUUAUUUUGGUCGGUUUUU .............................................(((.((((...((((((((........)))))))))))).)))..... ( -9.24 = -9.13 + -0.11)

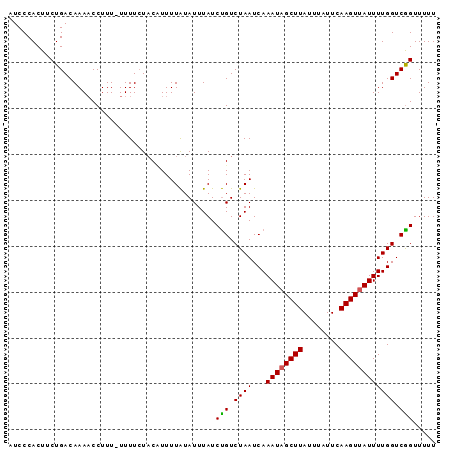
| Location | 8,039,545 – 8,039,644 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8039545 99 - 23771897 UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU-UUUUAUCCCUUUUAUAUUUAUCUGUCUAAUCAAAUA ((((.......))))((((((...))))))(((((.......)))))....((((........-......................)))).......... ( -18.31) >DroSec_CAF1 10759 99 - 1 UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGACAUUU-UUUUAUCCUUUUUAUAUUUAUCUAUCUAAUCAAAUA ...(((...(((...((((((...))))))(((((.......)))))....)))..)))....-.................................... ( -19.10) >DroSim_CAF1 10741 98 - 1 UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGAC-UUU-UUUUAUCCUUUUUAUAUUUAUCUAUCUAAUCAAAUA ...(((...(((...((((((...))))))(((((.......)))))....)))..)))-...-.................................... ( -18.60) >DroEre_CAF1 10572 99 - 1 UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU-UUUUCUACAUUUUGUAUUUAUCCGUCUAAUCAAAUA .(((.(((((.....((((((...))))))(((((.......))))).)))))..........-.....(((.....)))......)))........... ( -18.70) >DroYak_CAF1 11005 100 - 1 UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAUCCUUUUUUUUCUACUUUUUAUAUUUAUCUGUCUAAUCAAAUU ((((.......))))((((((...))))))(((((.......)))))....((((...............................)))).......... ( -18.28) >consensus UGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU_UUUUAUCCUUUUUAUAUUUAUCUGUCUAAUCAAAUA ...((.((((.....((((((...))))))(((((.......))))).)))))).............................................. (-18.00 = -18.00 + 0.00)

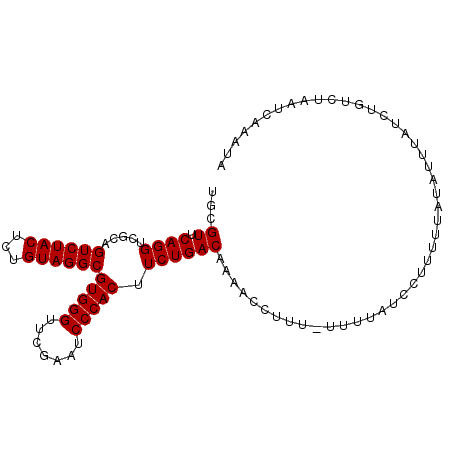
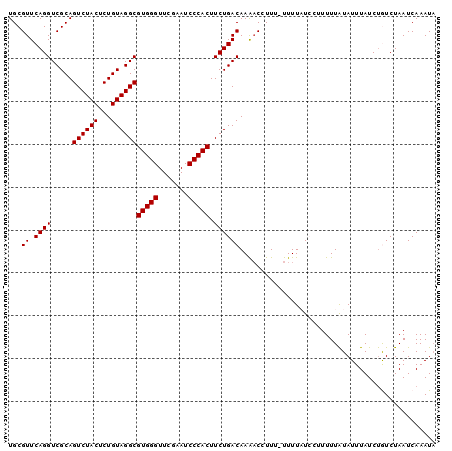
| Location | 8,039,569 – 8,039,684 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8039569 115 - 23771897 GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU-UUUUAUCCCUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......)))))))))))).........-............ ( -36.10) >DroSec_CAF1 10783 115 - 1 GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGACAUUU-UUUUAUCCUUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......)))))))))))).........-............ ( -36.10) >DroSim_CAF1 10765 114 - 1 GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGAC-UUU-UUUUAUCCUUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......)))))))))))).....-...-............ ( -36.10) >DroEre_CAF1 10596 115 - 1 GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU-UUUUCUACAUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......)))))))))))).........-............ ( -36.10) >DroYak_CAF1 11029 116 - 1 GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAUCCUUUUUUUUCUACUUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))...................... ( -36.10) >consensus GUUACUUUCAUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAAACCUUU_UUUUAUCCUUUU ............(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))...................... (-36.10 = -36.10 + 0.00)

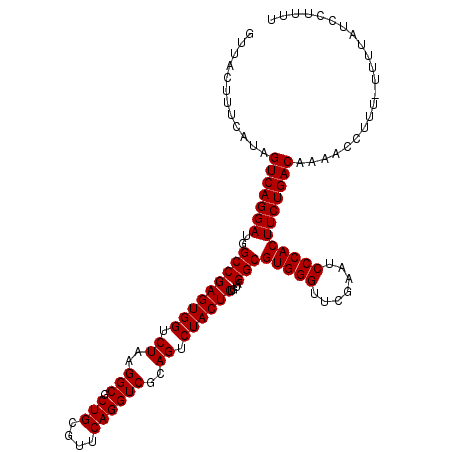
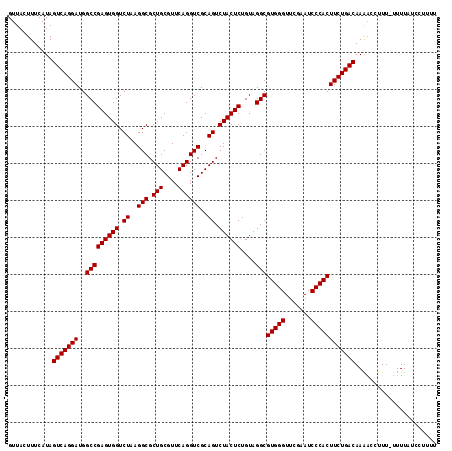
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:25 2006